4Y70
 
 | | Yeast 20S proteasome in complex with Ac-LAV-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAV-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y71
 
 | | Factor Xa complex with GTC000398 | | Descriptor: | 6-chloro-N-{(3S)-1-[(2S)-1-(4-methyl-5-oxo-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}naphthalene-2-sulf onamide, CALCIUM ION, Coagulation factor X | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Factor Xa complex with GTC000398
To be Published
|
|
4Y72
 
 | | Human CDK1/CyclinB1/CKS2 With Inhibitor | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Brown, N.R, Korolchuk, S, Martin, M.P, Stanley, W, Moukhametzianov, R, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CDK1 structures reveal conserved and unique features of the essential cell cycle CDK.
Nat Commun, 6, 2015
|
|
4Y73
 
 | | Crystal structure of IRAK4 kinase domain with inhibitor | | Descriptor: | 5-{[(1R,2S)-2-aminocyclohexyl]amino}-N-[1-methyl-3-(trifluoromethyl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Interleukin-1 receptor-associated kinase 4 | | Authors: | Lesburg, C.A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of 5-Amino-N-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide Inhibitors of IRAK4.
Acs Med.Chem.Lett., 6, 2015
|
|
4Y74
 
 | | Yeast 20S proteasome in complex with Ac-LAL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAL-ep, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y75
 
 | | Yeast 20S proteasome in complex with Ac-PAF-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PAF-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y76
 
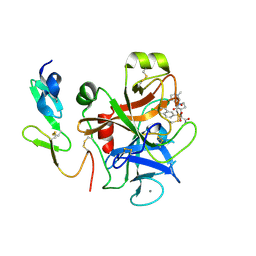 | | Factor Xa complex with GTC000401 | | Descriptor: | CALCIUM ION, Coagulation factor X, N~2~-[(6-chloronaphthalen-2-yl)sulfonyl]-N~2~-{(3S)-1-[(2S)-1-(4-methyl-1,4-diazepan-1-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}glycinamide | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4Y77
 
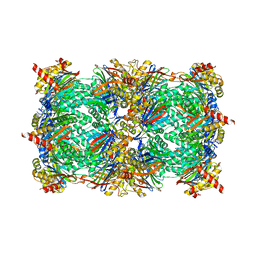 | | Yeast 20S proteasome in complex with Ac-LAF-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAF-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y78
 
 | | Yeast 20S proteasome in complex with Ac-LAD-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-LAD-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
4Y79
 
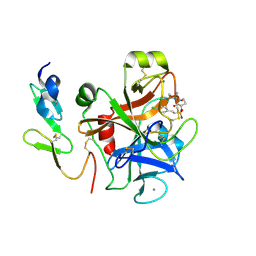 | | Factor Xa complex with GTC000406 | | Descriptor: | (E)-2-(4-chlorophenyl)-N-{(3S)-1-[(2S)-1-(morpholin-4-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}ethenesulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
4Y7A
 
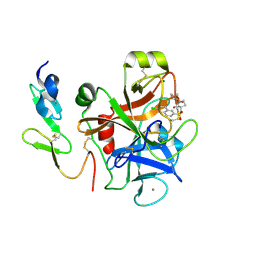 | | Factor Xa complex with GTC000422 | | Descriptor: | CALCIUM ION, Coagulation factor X, MAGNESIUM ION, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Factor Xa complex with GTC000422
to be published
|
|
4Y7B
 
 | | Factor Xa complex with GTC000441 | | Descriptor: | 6-chloro-N-{(3S)-1-[(2S)-1-{(1R,5S)-7-[2-(methylamino)ethyl]-3,7-diazabicyclo[3.3.1]non-3-yl}-1-oxopropan-2-yl]-2-oxopy rrolidin-3-yl}naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Factor Xa complex with GTC000441
to be published
|
|
4Y7C
 
 | | rat CYPOR mutant - G141del/E142N | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-13 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
4Y7D
 
 | | Alpha/beta hydrolase fold protein from Nakamurella multipartita | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION, SODIUM ION | | Authors: | Cuff, M.E, OSIPIUK, J, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Alpha/beta hydrolase fold protein from Nakamurella multipartita.
to be published
|
|
4Y7E
 
 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus with mannohexaose | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2015-02-14 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
4Y7F
 
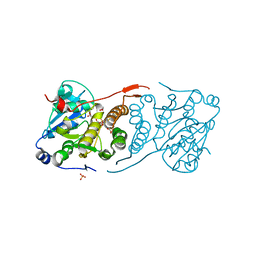 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and 3-(phosphonooxy)propanoic acid (PPA) - GpgS Mn2+ UDP-Glc PPA | | Descriptor: | 1,2-ETHANEDIOL, 3-(phosphonooxy)propanoic acid, Glucosyl-3-phosphoglycerate synthase, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.231 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4Y7G
 
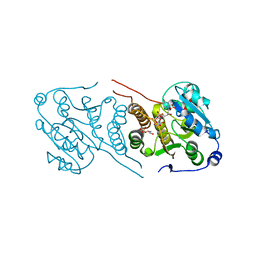 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and glycerol 3-phosphate (G3P) - GpgS Mn2+ UDP-Glc G3P | | Descriptor: | Glucosyl-3-phosphoglycerate synthase, MANGANESE (II) ION, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4Y7I
 
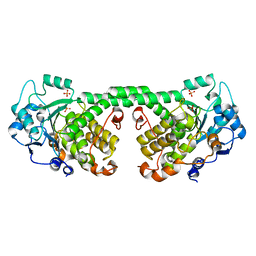 | | Crystal Structure of MTMR8 | | Descriptor: | Myotubularin-related protein 8, PHOSPHATE ION | | Authors: | Yoo, K, Lee, J, Son, J, Shin, W, Im, D, Heo, Y.S. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the catalytic phosphatase domain of MTMR8: implications for dimerization, membrane association and reversible oxidation.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Y7J
 
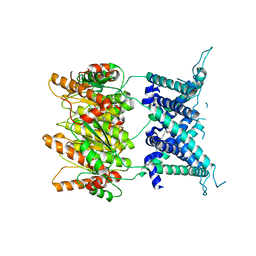 | | Structure of an archaeal mechanosensitive channel in expanded state | | Descriptor: | Large conductance mechanosensitive channel protein,Riboflavin synthase, nonyl beta-D-glucopyranoside | | Authors: | Li, J, Liu, Z. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Mechanical coupling of the multiple structural elements of the large-conductance mechanosensitive channel during expansion
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4Y7K
 
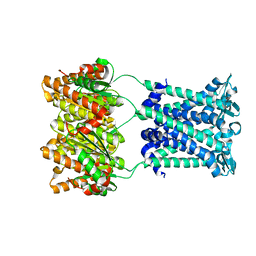 | | Structure of an archaeal mechanosensitive channel in closed state | | Descriptor: | Large conductance mechanosensitive channel protein,Riboflavin synthase | | Authors: | Li, J, Liu, Z. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Mechanical coupling of the multiple structural elements of the large-conductance mechanosensitive channel during expansion
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4Y7L
 
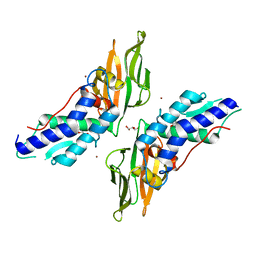 | | T6SS protein TssM C-terminal domain (869-1107) from EAEC | | Descriptor: | GLYCEROL, Type VI secretion protein IcmF, ZINC ION | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
4Y7M
 
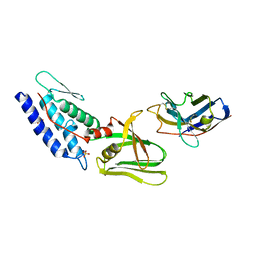 | | T6SS protein TssM C-terminal domain (835-1129) from EAEC | | Descriptor: | Hi113 protein, SULFATE ION, Type VI secretion protein IcmF | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
4Y7N
 
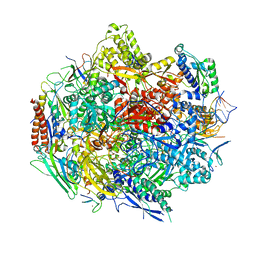 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4Y7O
 
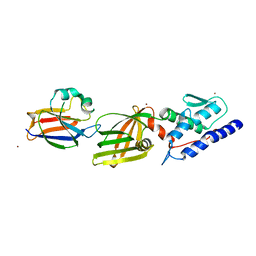 | | T6SS protein TssM C-terminal domain (869-1107) from EAEC | | Descriptor: | Type VI secretion protein IcmF, Type VI secretion system protein VasD, ZINC ION | | Authors: | Nguyen, V.S, Spinelli, S, Durand, E, Roussel, A, Cambillau, C. | | Deposit date: | 2015-02-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Biogenesis and structure of a type VI secretion membrane core complex.
Nature, 523, 2015
|
|
4Y7P
 
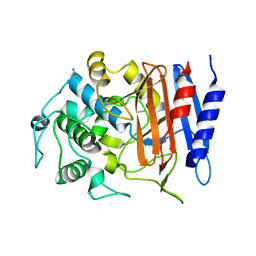 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
