1KTZ
 
 | | Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3 | | Descriptor: | TGF-beta Type II Receptor, TRANSFORMING GROWTH FACTOR BETA 3 | | Authors: | Hart, P.J, Deep, S, Taylor, A.B, Shu, Z, Hinck, C.S, Hinck, A.P. | | Deposit date: | 2002-01-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex.
Nat.Struct.Biol., 9, 2002
|
|
1KU0
 
 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
1KU1
 
 | | Crystal Structure of the Sec7 Domain of Yeast GEA2 | | Descriptor: | ARF guanine-nucleotide exchange factor 2 | | Authors: | Renault, L, Christova, P, Guibert, B, Pasqualato, S, Cherfils, J. | | Deposit date: | 2002-01-20 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of domain closure of Sec7 domains and role in BFA sensitivity.
Biochemistry, 41, 2002
|
|
1KU5
 
 | | Crystal Structure of recombinant histone HPhA from hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, HPhA, SULFATE ION | | Authors: | Li, T, Sun, F, Ji, X, Feng, Y, Rao, Z. | | Deposit date: | 2002-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii
J.MOL.BIOL., 325, 2003
|
|
1KU6
 
 | | Fasciculin 2-Mouse Acetylcholinesterase Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Burmeister, W, Taylor, P, Marchot, P. | | Deposit date: | 2002-01-21 | | Release date: | 2003-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site.
EMBO J., 22, 2003
|
|
1KU8
 
 | | Crystal structure of Jacalin | | Descriptor: | JACALIN ALPHA CHAIN, JACALIN BETA CHAIN | | Authors: | Bourne, Y, Astoul, C.H, Zamboni, V, Peumans, W.J, Menu-Bouaouiche, L, Van Damme, E.J.M, Barre, A, Rouge, P. | | Deposit date: | 2002-01-21 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose.
Biochem.J., 364, 2002
|
|
1KUF
 
 | | High-resolution Crystal Structure of a Snake Venom Metalloproteinase from Taiwan Habu | | Descriptor: | CADMIUM ION, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Yuann, J.M, Wang, A.H.J. | | Deposit date: | 2002-01-21 | | Release date: | 2002-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The 1.35 A structure of cadmium-substituted TM-3, a snake-venom metalloproteinase from Taiwan habu: elucidation of a TNFalpha-converting enzyme-like active-site structure with a distorted octahedral geometry of cadmium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KUG
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with its endogenous inhibitor pENW | | Descriptor: | CADMIUM ION, ENW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUH
 
 | | ZINC PROTEASE FROM STREPTOMYCES CAESPITOSUS | | Descriptor: | CALCIUM ION, ZINC ION, ZINC PROTEASE | | Authors: | Kurisu, G, Kinoshita, T, Sugimoto, A, Nagara, A, Kai, Y, Kasai, N, Harada, S. | | Deposit date: | 1996-02-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the zinc endoprotease from Streptomyces caespitosus.
J.Biochem.(Tokyo), 121, 1997
|
|
1KUI
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEQW. | | Descriptor: | CADMIUM ION, EQW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUJ
 
 | | Crystal structure of Jacalin complexed with 1-O-methyl-alpha-D-mannose | | Descriptor: | JACALIN ALPHA CHAIN, JACALIN BETA CHAIN, methyl alpha-D-mannopyranoside | | Authors: | Bourne, Y, Astoul, C.H, Zamboni, V, Peumans, W.J, Menu-Bouaouiche, L, Van Damme, E.J.M, Barre, A, Rouge, P. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose.
Biochem.J., 364, 2002
|
|
1KUK
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEKW. | | Descriptor: | CADMIUM ION, EKW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUQ
 
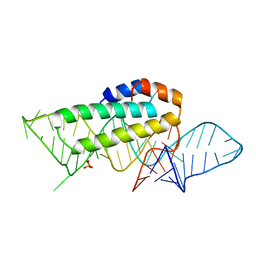 | | CRYSTAL STRUCTURE OF T3C MUTANT S15 RIBOSOMAL PROTEIN IN COMPLEX WITH 16S RRNA | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, SULFATE ION | | Authors: | Nikulin, A.D, Tishchenko, S, Revtovich, S, Ehresmann, B, Ehresmann, C, Dumas, P, Garber, M, Nikonov, S, Nevskaya, N. | | Deposit date: | 2002-01-22 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Role of N-terminal helix in interaction of ribosomal protein S15 with 16S rRNA.
Biochemistry Mosc., 69, 2004
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1KV0
 
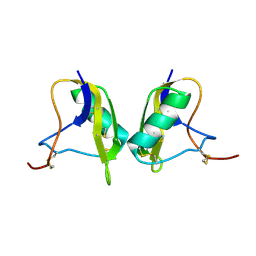 | | Cis/trans Isomerization of Non-prolyl Peptide Bond Observed in Crystal Structure of an Scorpion Toxin | | Descriptor: | Alpha-like toxin BmK-M7 | | Authors: | Guan, R.J, He, X.L, Wang, M, Xiang, Y, Wang, D.C. | | Deposit date: | 2002-01-23 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism governing cis and trans isomeric states and an intramolecular switch for cis/trans isomerization of a non-proline peptide bond observed in crystal structures of scorpion toxins.
J.Mol.Biol., 341, 2004
|
|
1KV3
 
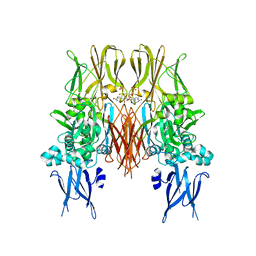 | | HUMAN TISSUE TRANSGLUTAMINASE IN GDP BOUND FORM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Protein-glutamine gamma-glutamyltransferase | | Authors: | Liu, S, Cerione, R.A, Clardy, J. | | Deposit date: | 2002-01-24 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the guanine nucleotide-binding activity of tissue transglutaminase and its regulation of transamidation activity.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KV5
 
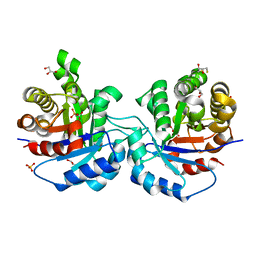 | | Structure of Trypanosoma brucei brucei TIM with the salt-bridge-forming residue Arg191 mutated to Ser | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PHOSPHOGLYCOLIC ACID, GLYCEROL, ... | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-01-25 | | Release date: | 2002-03-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The importance of the conserved Arg191-Asp227 salt bridge of triosephosphate isomerase for folding, stability, and catalysis
FEBS Lett., 518, 2002
|
|
1KV6
 
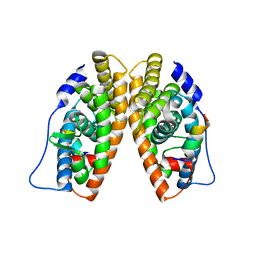 | | X-ray structure of the orphan nuclear receptor ERR3 ligand-binding domain in the constitutively active conformation | | Descriptor: | ESTROGEN-RELATED RECEPTOR GAMMA, steroid receptor coactivator 1 | | Authors: | Greschik, H, Wurtz, J.-M, Sanglier, S, Bourguet, W, van Dorsselaer, A, Moras, D, Renaud, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-01-25 | | Release date: | 2003-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Evidence for Ligand-Independent Transcriptional Activation by the Estrogen-Related Receptor 3
Mol.Cell, 9, 2002
|
|
1KV7
 
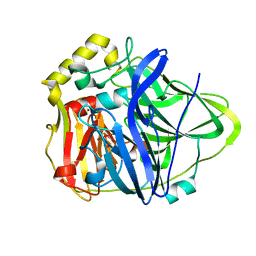 | | Crystal Structure of CueO, a multi-copper oxidase from E. coli involved in copper homeostasis | | Descriptor: | COPPER (II) ION, CU-O-CU LINKAGE, PROBABLE BLUE-COPPER PROTEIN YACK | | Authors: | Roberts, S.A, Weichsel, A, Grass, G, Thakali, K, Hazzard, J.T, Tollin, G, Rensing, C, Montfort, W.R. | | Deposit date: | 2002-01-25 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KV8
 
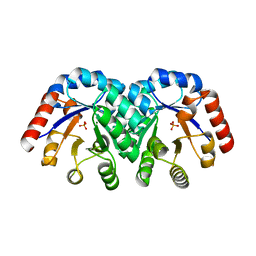 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
1KV9
 
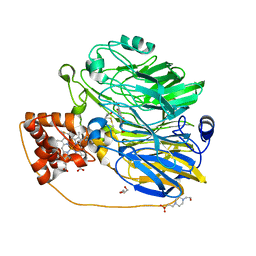 | | Structure at 1.9 A Resolution of a Quinohemoprotein Alcohol Dehydrogenase from Pseudomonas putida HK5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETONE, CALCIUM ION, ... | | Authors: | Chen, Z.-W, Matsushita, K, Yamashita, T, Fujii, T, Toyama, H, Adachi, O, Bellamy, H.D, Mathews, F.S. | | Deposit date: | 2002-01-25 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.9 A resolution of a quinohemoprotein alcohol dehydrogenase from Pseudomonas putida HK5.
Structure, 10, 2002
|
|
1KVA
 
 | | E. COLI RIBONUCLEASE HI D134A MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVB
 
 | | E. COLI RIBONUCLEASE HI D134H MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVC
 
 | | E. COLI RIBONUCLEASE HI D134N MUTANT | | Descriptor: | RIBONUCLEASE H | | Authors: | Kashiwagi, T, Jeanteur, D, Haruki, M, Katayanagi, K, Kanaya, S, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI.
Protein Eng., 9, 1996
|
|
1KVD
 
 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN, SULFATE ION | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
