1K3O
 
 | | Crystal Structure Analysis of apo Glutathione S-Transferase | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1 | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
1K3R
 
 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | Descriptor: | conserved protein MT0001 | | Authors: | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-03 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
1K3S
 
 | | Type III Secretion Chaperone SigE | | Descriptor: | PHOSPHATE ION, SigE | | Authors: | Bertero, M.G, Luo, Y, Frey, E.A, Pfuetzner, R.A, Wenk, M.R, Creagh, L, Marcus, S.L, Lim, D, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2001-10-03 | | Release date: | 2001-11-28 | | Last modified: | 2016-05-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biochemical characterization of the type III secretion chaperones CesT and SigE.
Nat.Struct.Biol., 8, 2001
|
|
1K3T
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi Complexed with Chalepin, a Coumarin Derivative Inhibitor | | Descriptor: | 6-(1,1-DIMETHYLALLYL)-2-(1-HYDROXY-1-METHYLETHYL)-2,3-DIHYDRO-7H-FURO[3,2-G]CHROMEN-7-ONE, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pavao, F. | | Deposit date: | 2001-10-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase complexed with chalepin, a natural product inhibitor, at 1.95 A resolution.
FEBS Lett., 520, 2002
|
|
1K3U
 
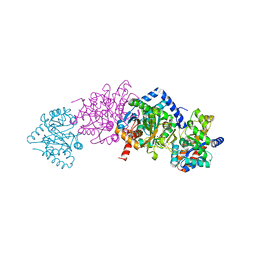 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-04 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K3Y
 
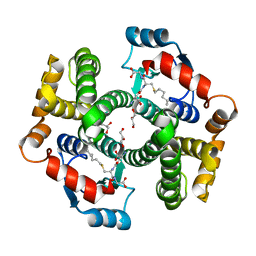 | | Crystal Structure Analysis of human Glutathione S-transferase with S-hexyl glutatione and glycerol at 1.3 Angstrom | | Descriptor: | GLUTATHIONE S-TRANSFERASE A1, GLYCEROL, S-HEXYLGLUTATHIONE | | Authors: | Le Trong, I, Stenkamp, R.E, Ibarra, C, Atkins, W.M, Adman, E.T. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3-A resolution structure of human glutathione S-transferase with S-hexyl glutathione bound reveals possible extended ligandin binding site.
Proteins, 48, 2002
|
|
1K40
 
 | |
1K44
 
 | |
1K46
 
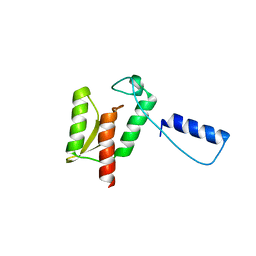 | | Crystal Structure of the Type III Secretory Domain of Yersinia YopH Reveals a Domain-Swapped Dimer | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH | | Authors: | Smith, C.L, Khandelwal, P, Keliikuli, K, Zuiderweg, E.R.P, Saper, M.A. | | Deposit date: | 2001-10-05 | | Release date: | 2001-11-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the type III secretion and substrate-binding domain of Yersinia YopH phosphatase.
Mol.Microbiol., 42, 2001
|
|
1K47
 
 | |
1K4C
 
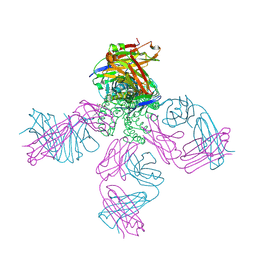 | | Potassium Channel KcsA-Fab complex in high concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
1K4D
 
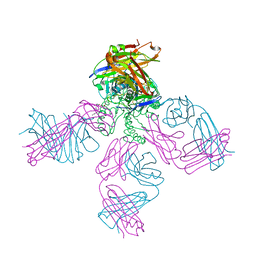 | | Potassium Channel KcsA-Fab complex in low concentration of K+ | | Descriptor: | DIACYL GLYCEROL, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Zhou, Y, Morais-Cabral, J.H, Kaufman, A, MacKinnon, R. | | Deposit date: | 2001-10-07 | | Release date: | 2001-11-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution.
Nature, 414, 2001
|
|
1K4E
 
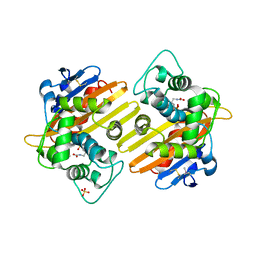 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASES OXA-10 DETERMINED BY MAD PHASING WITH SELENOMETHIONINE | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | STRUCTURE OF CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
1K4F
 
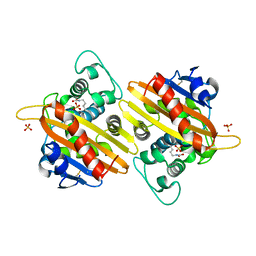 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 AT 1.6 A RESOLUTION | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Fonze, E, Bouillene, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-08 | | Release date: | 2001-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the class D beta-lactamase OXA-2
To be Published
|
|
1K4G
 
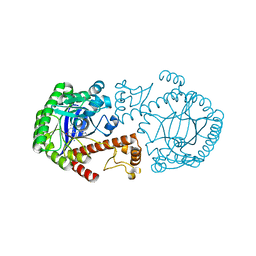 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
ChemBioChem, 3, 2002
|
|
1K4H
 
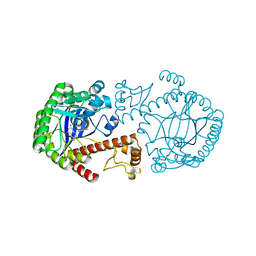 | | CRYSTAL STRUCTURE OF TRNA-GUANINE TRANSGLYCOSYLASE (TGT) COMPLEXED WITH 2,6-Diamino-8-propylsulfanylmethyl-3H-quinazoline-4-one | | Descriptor: | 2,6-DIAMINO-8-PROPYLSULFANYLMETHYL-3H-QUINAZOLINE-4-ONE, TRNA-GUANINE-TRANSGLYCOSYLASE, ZINC ION | | Authors: | Brenk, R, Meyer, E.A, Castellano, R.K, Furler, M, Stubbs, M.T, Klebe, G, Diederich, F. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design, synthesis, and in vitro evaluation of inhibitors for prokaryotic tRNA-guanine transglycosylase: a dramatic sulfur effect on binding affinity.
Chembiochem, 3, 2002
|
|
1K4Q
 
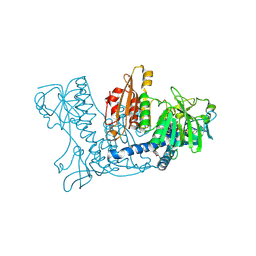 | | Human Glutathione Reductase Inactivated by Peroxynitrite | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutathione Reductase | | Authors: | Savvides, S.N, Scheiwein, M, Boehme, C.C, Arteel, G.E, Karplus, P.A, Becker, K, Schirmer, R.H. | | Deposit date: | 2001-10-08 | | Release date: | 2002-01-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the antioxidant enzyme glutathione reductase inactivated by peroxynitrite.
J.Biol.Chem., 277, 2002
|
|
1K4T
 
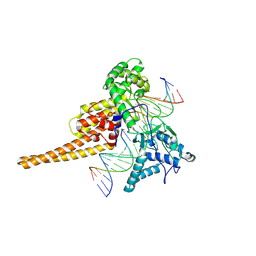 | | HUMAN DNA TOPOISOMERASE I (70 KDA) IN COMPLEX WITH THE POISON TOPOTECAN AND COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, 2-(1-DIMETHYLAMINOMETHYL-2-HYDROXY-8-HYDROXYMETHYL-9-OXO-9,11-DIHYDRO-INDOLIZINO[1,2-B]QUINOLIN-7-YL)-2-HYDROXY-BUTYRIC ACID, 5'-D(*(TGP)P*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Staker, B.L, Hjerrild, K, Feese, M.D, Behnke, C.A, Burgin Jr, A.B, Stewart, L.J. | | Deposit date: | 2001-10-08 | | Release date: | 2002-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of topoisomerase I poisoning by a camptothecin analog
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K4W
 
 | | X-ray structure of the orphan nuclear receptor ROR beta ligand-binding domain in the active conformation | | Descriptor: | Nuclear receptor ROR-beta, STEARIC ACID, steroid receptor coactivator-1 | | Authors: | Stehlin, C, Wurtz, J.M, Steinmetz, A, Greiner, E, Schuele, R, Moras, D, Renaud, J.P. | | Deposit date: | 2001-10-09 | | Release date: | 2002-04-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation.
EMBO J., 20, 2001
|
|
1K50
 
 | | A V49A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K51
 
 | | A G55A Mutation Induces 3D Domain Swapping in the B1 Domain of Protein L from Peptostreptococcus magnus | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K52
 
 | | Monomeric Protein L B1 Domain with a K54G mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K53
 
 | | Monomeric Protein L B1 Domain with a G15A Mutation | | Descriptor: | Protein L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Johnsen, K, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-10-09 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-site mutations induce 3D domain swapping in the B1 domain of protein L from Peptostreptococcus magnus.
Structure, 9, 2001
|
|
1K54
 
 | | OXA-10 class D beta-lactamase partially acylated with reacted 6beta-(1-hydroxy-1-methylethyl) penicillanic acid | | Descriptor: | (1R)-2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, Beta lactamase OXA-10, ... | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K55
 
 | | OXA 10 class D beta-lactamase at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Beta lactamase OXA-10, SULFATE ION | | Authors: | Golemi, D, Maveyraud, L, Vakulenko, S, Samama, J.P, Mobashery, S. | | Deposit date: | 2001-10-10 | | Release date: | 2001-11-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Critical involvement of a carbamylated lysine in catalytic function of class D beta-lactamases.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
