1DSS
 
 | |
1DST
 
 | |
1DSU
 
 | |
1DT1
 
 | | THERMUS THERMOPHILUS CYTOCHROME C552 SYNTHESIZED BY ESCHERICHIA COLI | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Fee, J.A, Chen, Y, Hill, M.J, Gomez-Moran, E, Loehr, T, Ai, J, Thony-Meyer, L, Williams, P.A, Stura, E, Sridhar, V, McRee, D.E. | | Deposit date: | 2000-01-10 | | Release date: | 2000-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrity of thermus thermophilus cytochrome c552 synthesized by Escherichia coli cells expressing the host-specific cytochrome c maturation genes, ccmABCDEFGH: biochemical, spectral, and structural characterization of the recombinant protein.
Protein Sci., 9, 2000
|
|
1DT4
 
 | | CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 1 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L. | | Deposit date: | 2000-01-11 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1DT6
 
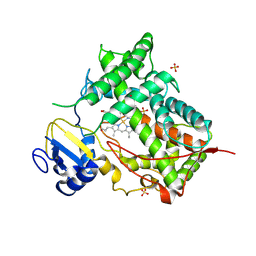 | | STRUCTURE OF MAMMALIAN CYTOCHROME P450 2C5 | | Descriptor: | CYTOCHROME P450 2C5, PROTOPORPHYRIN IX CONTAINING FE, SAMARIUM (III) ION, ... | | Authors: | Williams, P.A, Cosme, J, Sridhar, V, Johnson, E.F, McRee, D.E. | | Deposit date: | 2000-01-11 | | Release date: | 2000-09-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity.
Mol.Cell, 5, 2000
|
|
1DT9
 
 | |
1DTG
 
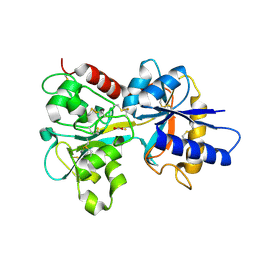 | | HUMAN TRANSFERRIN N-LOBE MUTANT H249E | | Descriptor: | CARBONATE ION, FE (III) ION, TRANSFERRIN | | Authors: | MacGillivray, R.T, Bewley, M.C, Smith, C.A, He, Q.Y, Mason, A.B. | | Deposit date: | 2000-01-12 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutation of the iron ligand His 249 to Glu in the N-lobe of human transferrin abolishes the dilysine "trigger" but does not significantly affect iron release.
Biochemistry, 39, 2000
|
|
1DTI
 
 | |
1DTJ
 
 | | CRYSTAL STRUCTURE OF NOVA-2 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | RNA-BINDING NEUROONCOLOGICAL VENTRAL ANTIGEN 2 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L, Musunuru, K, Zhong, R, Darnell, R.B, Burley, S.K. | | Deposit date: | 2000-01-12 | | Release date: | 2000-02-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
1DTM
 
 | |
1DTO
 
 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
1DTP
 
 | |
1DTQ
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTT
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-2 (PETT130A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-CHLORO-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-04-02 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
1DTU
 
 | | BACILLUS CIRCULANS STRAIN 251 CYCLODEXTRIN GLYCOSYLTRANSFERASE: A MUTANT Y89D/S146P COMPLEXED TO AN HEXASACCHARIDE INHIBITOR | | Descriptor: | 1-AMINO-2,3-DIHYDROXY-5-HYDROXYMETHYL CYCLOHEX-5-ENE, CALCIUM ION, PROTEIN (CYCLODEXTRIN GLYCOSYLTRANSFERASE), ... | | Authors: | Uitdehaag, J.C.M, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Rational design of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 to increase alpha-cyclodextrin production.
J.Mol.Biol., 296, 2000
|
|
1DTX
 
 | |
1DU5
 
 | | THE CRYSTAL STRUCTURE OF ZEAMATIN. | | Descriptor: | ZEAMATIN | | Authors: | Batalia, M.A, Monzingo, A.F, Ernst, S, Roberts, W, Robertus, J.D. | | Deposit date: | 2000-01-14 | | Release date: | 2000-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the antifungal protein zeamatin, a member of the thaumatin-like, PR-5 protein family.
Nat.Struct.Biol., 3, 1996
|
|
1DUB
 
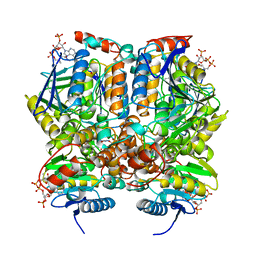 | | 2-ENOYL-COA HYDRATASE, DATA COLLECTED AT 100 K, PH 6.5 | | Descriptor: | 2-ENOYL-COA HYDRATASE, ACETOACETYL-COENZYME A | | Authors: | Wierenga, R.K, Engel, C.K. | | Deposit date: | 1996-06-10 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket.
EMBO J., 15, 1996
|
|
1DUC
 
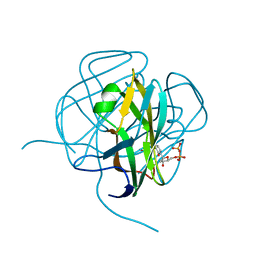 | | EIAV DUTPASE DUDP/STRONTIUM COMPLEX | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-DIPHOSPHATE, STRONTIUM ION | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1DUD
 
 | |
1DUG
 
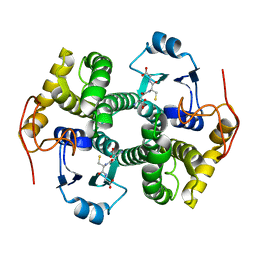 | | STRUCTURE OF THE FIBRINOGEN G CHAIN INTEGRIN BINDING AND FACTOR XIIIA CROSSLINKING SITES OBTAINED THROUGH CARRIER PROTEIN DRIVEN CRYSTALLIZATION | | Descriptor: | GLUTATHIONE, chimera of GLUTATHIONE S-TRANSFERASE-synthetic LINKEr-C-TERMINAL FIBRINOGEN GAMMA CHAIN | | Authors: | Ware, S, Donahue, J.P, Hawiger, J, Anderson, W.F. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the fibrinogen gamma-chain integrin binding and factor XIIIa cross-linking sites obtained through carrier protein driven crystallization.
Protein Sci., 8, 1999
|
|
1DUH
 
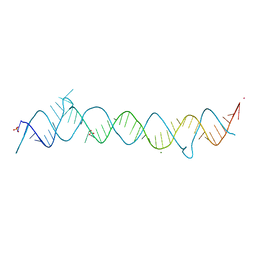 | | CRYSTAL STRUCTURE OF THE CONSERVED DOMAIN IV OF E. COLI 4.5S RNA | | Descriptor: | 4.5S RNA DOMAIN IV, LUTETIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Jovine, L, Hainzl, T, Oubridge, C, Scott, W.G, Li, J, Sixma, T.K, Wonacott, A, Skarzynski, T, Nagai, K. | | Deposit date: | 2000-01-17 | | Release date: | 2000-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA.
Structure Fold.Des., 8, 2000
|
|
1DUI
 
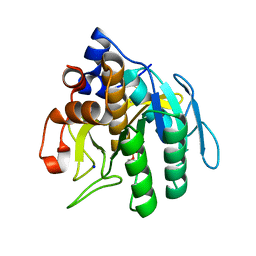 | | Subtilisin BPN' from Bacillus amyloliquefaciens, crystal growth mutant | | Descriptor: | DIISOPROPYL PHOSPHONATE, PROTEIN (SUBTILISIN BPN'), SODIUM ION | | Authors: | Pan, Q, Gallagher, D.T. | | Deposit date: | 2000-01-17 | | Release date: | 2000-01-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing Protein Interaction Chemistry Through Crystal Growth: Structure, Mutation, and Mechanism in Subtilisin s88
J.Cryst.Growth, 212, 2000
|
|
1DUK
 
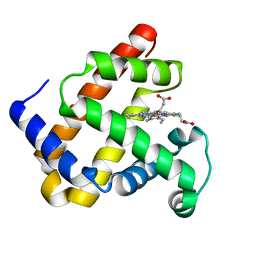 | | WILD-TYPE RECOMBINANT SPERM WHALE METAQUOMYOGLOBIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, WILD-TYPE RECOMBINANT SPERM WHALE METAQUOMYOGLOBIN | | Authors: | Barrick, D, Dahlquist, F.W. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Trans-substitution of the proximal hydrogen bond in myoglobin: I. Structural consequences of hydrogen bond deletion.
Proteins, 39, 2000
|
|
