1IGB
 
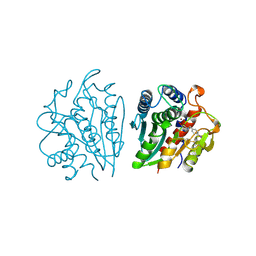 | | AEROMONAS PROTEOLYTICA AMINOPEPTIDASE COMPLEXED WITH THE INHIBITOR PARA-IODO-D-PHENYLALANINE HYDROXAMATE | | Descriptor: | AMINOPEPTIDASE, PARA-IODO-D-PHENYLALANINE HYDROXAMIC ACID, ZINC ION | | Authors: | Chevrier, B, D'Orchymont, H, Schalk, C, Tarnus, C, Moras, D. | | Deposit date: | 1996-02-27 | | Release date: | 1996-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the Aeromonas proteolytica aminopeptidase complexed with a hydroxamate inhibitor. Involvement in catalysis of Glu151 and two zinc ions of the co-catalytic unit.
Eur.J.Biochem., 237, 1996
|
|
1IGC
 
 | |
1IGD
 
 | |
1IGI
 
 | |
1IGJ
 
 | |
1IGN
 
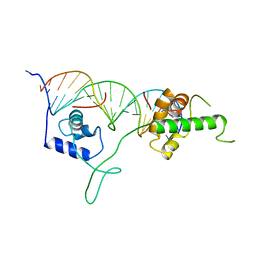 | | DNA-BINDING DOMAIN OF RAP1 IN COMPLEX WITH TELOMERIC DNA SITE | | Descriptor: | DNA (5'-D(*CP*CP*GP*CP*AP*CP*AP*CP*CP*CP*AP*CP*AP*CP*AP*CP*C P*AP*G)-3'), DNA (5'-D(*CP*CP*TP*GP*GP*TP*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G P*CP*G)-3'), PROTEIN (RAP1) | | Authors: | Koenig, P, Giraldo, R, Chapman, L, Rhodes, D. | | Deposit date: | 1996-02-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA.
Cell(Cambridge,Mass.), 85, 1996
|
|
1IGO
 
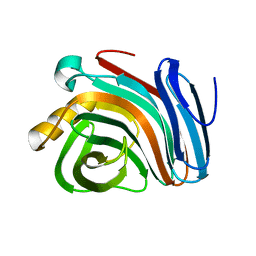 | | Family 11 xylanase | | Descriptor: | SULFATE ION, family 11 xylanase | | Authors: | Oakley, A.J, Thomson, C, Heinrich, T, Dunlop, R, Wilce, M.C.J. | | Deposit date: | 2001-04-18 | | Release date: | 2002-04-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of a family 11 xylanase from Bacillus subtillis B230 used for paper bleaching.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IGQ
 
 | |
1IGU
 
 | |
1IGV
 
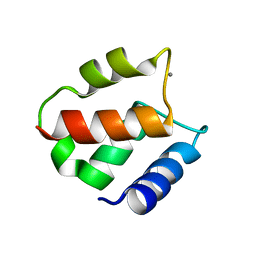 | | BOVINE CALBINDIN D9K BINDING MN2+ | | Descriptor: | MANGANESE (II) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1IGW
 
 | | Crystal Structure of the Isocitrate Lyase from the A219C mutant of Escherichia coli | | Descriptor: | Isocitrate lyase, MAGNESIUM ION, MERCURY (II) ION, ... | | Authors: | Britton, K.L, Abeysinghe, I.S.B, Baker, P.J, Barynin, V, Diehl, P, Langridge, S.J, McFadden, B.A, Sedelnikova, S.E, Stillman, T.J, Weeradechapon, K, Rice, D.W. | | Deposit date: | 2001-04-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure and domain organization of Escherichia coli isocitrate lyase.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IGX
 
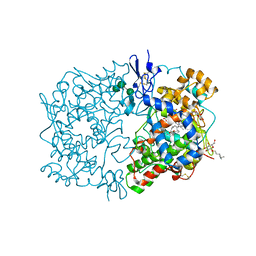 | | Crystal Structure of Eicosapentanoic Acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5,8,11,14,17-EICOSAPENTAENOIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
1IGZ
 
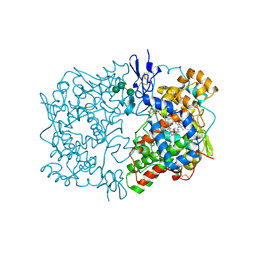 | | Crystal Structure of Linoleic acid Bound in the Cyclooxygenase Channel of Prostaglandin Endoperoxide H Synthase-1. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Malkowski, M.G, Thuresson, E.D, Smith, W.L, Garavito, R.M. | | Deposit date: | 2001-04-18 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of eicosapentaenoic and linoleic acids in the cyclooxygenase site of prostaglandin endoperoxide H synthase-1.
J.Biol.Chem., 276, 2001
|
|
1IH1
 
 | |
1IH2
 
 | |
1IH3
 
 | |
1IH4
 
 | |
1IH6
 
 | |
1IH7
 
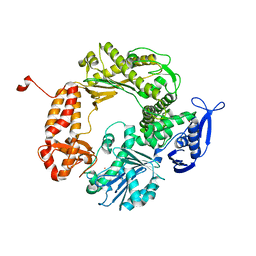 | | High-Resolution Structure of Apo RB69 DNA Polymerase | | Descriptor: | DNA POLYMERASE, GUANOSINE, POTASSIUM ION | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Replicating Complex of a Pol alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
1IH8
 
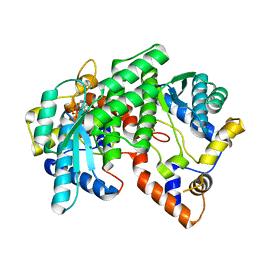 | | NH3-dependent NAD+ Synthetase from Bacillus subtilis Complexed with AMP-CPP and Mg2+ ions. | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NH(3)-DEPENDENT NAD(+) synthetase | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Jedrzejas, M, Brouillette, C, Brouillette, W, Muccio, D, Chattopadhyay, D, DeLucas, L. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IHA
 
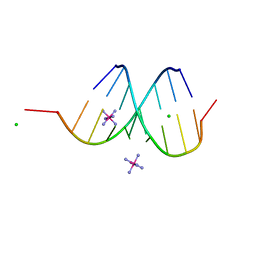 | | Structure of the Hybrid RNA/DNA R-GCUUCGGC-D[BR]U in Presence of RH(NH3)6+++ | | Descriptor: | 5'-R(*GP*CP*UP*UP*CP*GP*GP*C)-D(P*(BRU))-3', CHLORIDE ION, RHODIUM HEXAMINE ION | | Authors: | Cruse, W.B, Saludjian, P, Neuman, A, Prange, T. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Destabilizing effect of a fluorouracil extra base in a hybrid RNA duplex compared with bromo and chloro analogues.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IHB
 
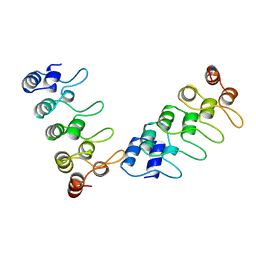 | | CRYSTAL STRUCTURE OF P18-INK4C(INK6) | | Descriptor: | CYCLIN-DEPENDENT KINASE 6 INHIBITOR | | Authors: | Ravichandran, V, Swaminathan, K, Marmorstein, R. | | Deposit date: | 1997-10-25 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the CDK4/6 inhibitory protein p18INK4c provides insights into ankyrin-like repeat structure/function and tumor-derived p16INK4 mutations.
Nat.Struct.Biol., 5, 1998
|
|
1IHC
 
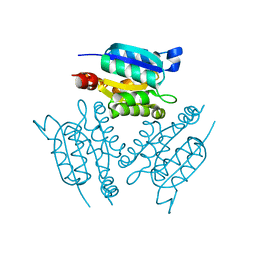 | | X-ray Structure of Gephyrin N-terminal Domain | | Descriptor: | Gephyrin | | Authors: | Sola, M, Kneussel, M, Heck, I.S, Betz, H, Weissenhorn, W. | | Deposit date: | 2001-04-21 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the trimeric N-terminal domain of gephyrin.
J.Biol.Chem., 276, 2001
|
|
1IHD
 
 | |
1IHF
 
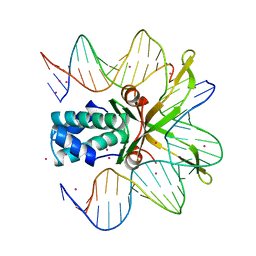 | | INTEGRATION HOST FACTOR/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (35-MER), DNA (5'-D(*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*C P*AP*CP*C)-3'), ... | | Authors: | Rice, P.A, Yang, S.-W, Mizuuchi, K, Nash, H.A. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn.
Cell(Cambridge,Mass.), 87, 1996
|
|
