1BZO
 
 | | THREE-DIMENSIONAL STRUCTURE OF PROKARYOTIC CU,ZN SUPEROXIDE DISMUTASE FROM P.LEIOGNATHI, SOLVED BY X-RAY CRYSTALLOGRAPHY. | | Descriptor: | COPPER (II) ION, PROTEIN (SUPEROXIDE DISMUTASE), URANYL (VI) ION, ... | | Authors: | Bordo, D, Matak, D, Djinovic-Carugo, K, Rosano, C, Pesce, A, Bolognesi, M, Stroppolo, M.E, Falconi, M, Battistoni, A, Desideri, A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase.
J.Mol.Biol., 285, 1999
|
|
1BZP
 
 | |
1BZQ
 
 | | COMPLEX OF A DROMEDARY SINGLE-DOMAIN VHH ANTIBODY FRAGMENT WITH RNASE A | | Descriptor: | PHOSPHATE ION, PROTEIN (ANTIBODY CAB-RN05), PROTEIN (RNASE A) | | Authors: | Decanniere, K, Desmyter, A, Gahroudhi, M, Lauwereys, M, Muyldermans, S, Wyns, L. | | Deposit date: | 1998-11-03 | | Release date: | 1998-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops.
Structure Fold.Des., 7, 1999
|
|
1BZR
 
 | | ATOMIC RESOLUTION CRYSTAL STRUCTURE ANALYSIS OF NATIVE DEOXY AND CO MYOGLOBIN FROM SPERM WHALE AT ROOM TEMPERATURE | | Descriptor: | CARBON MONOXIDE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kachalova, G.S, Popov, A.N, Bartunik, H.D. | | Deposit date: | 1998-11-03 | | Release date: | 1999-05-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A steric mechanism for inhibition of CO binding to heme proteins.
Science, 284, 1999
|
|
1BZS
 
 | | CRYSTAL STRUCTURE OF MMP8 COMPLEXED WITH HMR2909 | | Descriptor: | 2-(BIPHENYL-4-SULFONYL)-1,2,3,4-TETRAHYDRO-ISOQUINOLINE-3-CARBOXYLIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Schreuder, H, Brachvogel, V, Loenze, P. | | Deposit date: | 1998-11-04 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Quantitative structure-activity relationship of human neutrophil collagenase (MMP-8) inhibitors using comparative molecular field analysis and X-ray structure analysis.
J.Med.Chem., 42, 1999
|
|
1BZW
 
 | | PEANUT LECTIN COMPLEXED WITH C-LACTOSE | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PROTEIN (PEANUT LECTIN), ... | | Authors: | Ravishankar, R, Surolia, A, Vijayan, M, Lim, S, Kishi, Y. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Preferred Conformation of C-Lactose at the Free and Peanut Lectin Bound States
J.Am.Chem.Soc., 120, 1998
|
|
1BZX
 
 | | THE CRYSTAL STRUCTURE OF ANIONIC SALMON TRYPSIN IN COMPLEX WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | Descriptor: | CALCIUM ION, PROTEIN (BOVINE PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN) | | Authors: | Helland, R, Leiros, I, Berglund, G.I, Willassen, N.P, Smalas, A.O. | | Deposit date: | 1998-11-05 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of anionic salmon trypsin in complex with bovine pancreatic trypsin inhibitor.
Eur.J.Biochem., 256, 1998
|
|
1BZY
 
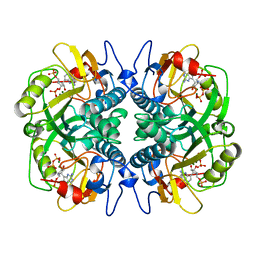 | | HUMAN HGPRTASE WITH TRANSITION STATE INHIBITOR | | Descriptor: | HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, MAGNESIUM ION, PHOSPHORIC ACID MONO-[5-(2-AMINO-4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-3,4-DIHYDROXY-PYRROLIDIN-2-YLMETHYL] ESTER, ... | | Authors: | Shi, W, Li, C, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 1998-11-05 | | Release date: | 1999-06-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human hypoxanthine-guanine phosphoribosyltransferase in complex with a transition-state analog inhibitor.
Nat.Struct.Biol., 6, 1999
|
|
1BZZ
 
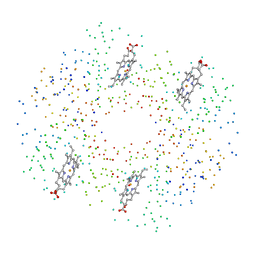 | | HEMOGLOBIN (ALPHA V1M) MUTANT | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kavanaugh, J.S, Arnone, A. | | Deposit date: | 1998-11-04 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and functional properties of human hemoglobins reassembled after synthesis in Escherichia coli.
Biochemistry, 38, 1999
|
|
1C02
 
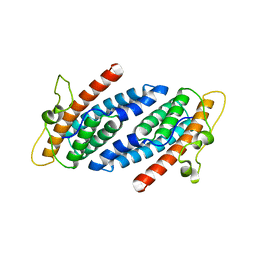 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | Descriptor: | PHOSPHOTRANSFERASE YPD1P | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C03
 
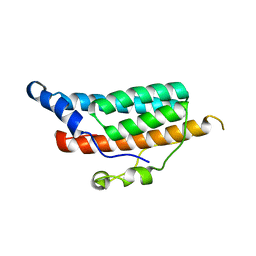 | | CRYSTAL STRUCTURE OF YPD1P (TRICLINIC FORM) | | Descriptor: | HYPOTHETICAL PROTEIN YDL235C | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C0A
 
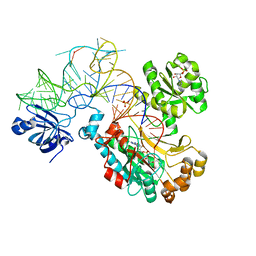 | | CRYSTAL STRUCTURE OF THE E. COLI ASPARTYL-TRNA SYNTHETASE : TRNAASP : ASPARTYL-ADENYLATE COMPLEX | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARTYL TRNA, ASPARTYL TRNA SYNTHETASE, ... | | Authors: | Eiler, S, Dock-Bregeon, A.-C, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-07-15 | | Release date: | 1999-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis of aspartyl-tRNA(Asp) in Escherichia coli--a snapshot of the second step.
EMBO J., 18, 1999
|
|
1C0B
 
 | |
1C0C
 
 | |
1C0I
 
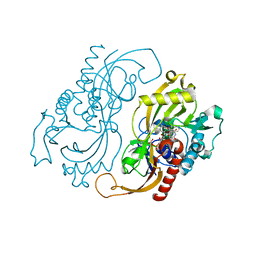 | | CRYSTAL STRUCTURE OF D-AMINO ACID OXIDASE IN COMPLEX WITH TWO ANTHRANYLATE MOLECULES | | Descriptor: | 2-AMINOBENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pollegioni, L, Diederichs, K, Molla, G, Umhau, S, Welte, W, Ghisla, S, Pilone, M.S. | | Deposit date: | 1999-07-16 | | Release date: | 2002-02-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Yeast d-amino Acid oxidase: structural basis of its catalytic properties
J.Mol.Biol., 324, 2002
|
|
1C0K
 
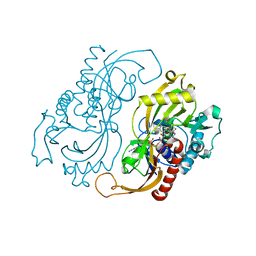 | | CRYSTAL STRUCTURE ANALYSIS OF D-AMINO ACID OXIDASE IN COMPLEX WITH L-LACTATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LACTIC ACID, PROTEIN (D-AMINO ACID OXIDASE) | | Authors: | Umhau, S, Molla, G, Diederichs, K, Pilone, M.S, Ghisla, S, Welte, W, Pollegioni, L. | | Deposit date: | 1999-07-16 | | Release date: | 2000-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C0L
 
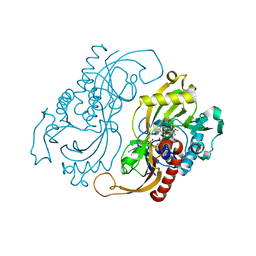 | | D-AMINO ACID OXIDASE: STRUCTURE OF SUBSTRATE COMPLEXES AT VERY HIGH RESOLUTION REVEAL THE CHEMICAL REACTTION MECHANISM OF FLAVIN DEHYDROGENATION | | Descriptor: | D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, TRIFLUOROALANINE | | Authors: | Umhau, S, Molla, G, Diederichs, K, Pilone, M.S, Ghisla, S, Welte, W. | | Deposit date: | 1999-07-16 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C0M
 
 | |
1C0N
 
 | | CSDB PROTEIN, NIFS HOMOLOGUE | | Descriptor: | ACETIC ACID, PROTEIN (CSDB PROTEIN), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Fujii, T, Maeda, M, Mihara, H, Kurihara, T, Esaki, N, Hata, Y. | | Deposit date: | 1999-07-17 | | Release date: | 2000-07-17 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a NifS homologue: X-ray structure analysis of CsdB, an Escherichia coli counterpart of mammalian selenocysteine lyase
Biochemistry, 39, 2000
|
|
1C0P
 
 | | D-AMINO ACIC OXIDASE IN COMPLEX WITH D-ALANINE AND A PARTIALLY OCCUPIED BIATOMIC SPECIES | | Descriptor: | D-ALANINE, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Umhau, S, Pollegioni, L, Molla, G, Diederichs, K, Welte, W, Pilone, S.M, Ghisla, S. | | Deposit date: | 1999-07-19 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The x-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1C0T
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+21.1326 | | Descriptor: | (R)-(+)9B-(3-METHYL)PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-5(9BH)-ONE, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1C0U
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH BM+50.0934 | | Descriptor: | (R)-(+) 5(9BH)-OXO-9B-PHENYL-2,3-DIHYDROTHIAZOLO[2,3-A]ISOINDOL-3-CARBOXYLIC ACID METHYL ESTER, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | Authors: | Ren, J, Esnouf, R.M, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 1999-07-19 | | Release date: | 2000-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystallographic analysis of the binding modes of thiazoloisoindolinone non-nucleoside inhibitors to HIV-1 reverse transcriptase and comparison with modeling studies.
J.Med.Chem., 42, 1999
|
|
1C0W
 
 | | CRYSTAL STRUCTURE OF THE COBALT-ACTIVATED DIPHTHERIA TOXIN REPRESSOR-DNA COMPLEX REVEALS A METAL BINDING SH-LIKE DOMAIN | | Descriptor: | COBALT (II) ION, DIPHTHERIA TOXIN REPRESSOR, DNA (5'-D(P*AP*TP*TP*AP*GP*GP*TP*TP*AP*GP*CP*CP*TP*AP*CP*CP*CP*TP*AP*AP*T)-3'), ... | | Authors: | Pohl, E, Holmes, R.K, Hol, W.G. | | Deposit date: | 1999-07-22 | | Release date: | 2000-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a cobalt-activated diphtheria toxin repressor-DNA complex reveals a metal-binding SH3-like domain.
J.Mol.Biol., 292, 1999
|
|
1C16
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE GAMMA/DELTA T CELL LIGAND T22 | | Descriptor: | MHC-LIKE PROTEIN T22, PROTEIN (BETA-2-MICROGLOBULIN) | | Authors: | Wingren, C, Crowley, M.P, Degano, M, Chien, Y, Wilson, I.A. | | Deposit date: | 1999-07-20 | | Release date: | 2000-01-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold.
Science, 287, 2000
|
|
1C1A
 
 | |
