1Q4X
 
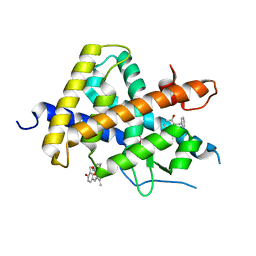 | | Crystal Structure of Human Thyroid Hormone Receptor beta LBD in complex with specific agonist GC-24 | | Descriptor: | Thyroid hormone receptor beta-1, [4-(3-BENZYL-4-HYDROXYBENZYL)-3,5-DIMETHYLPHENOXY]ACETIC ACID | | Authors: | Borngraeber, S, Budny, M.J, Chiellini, G, Cunha-Lima, S.T, Togashi, M, Webb, P, Baxter, J.D, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 2003-08-04 | | Release date: | 2004-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ligand selectivity by seeking hydrophobicity in thyroid hormone receptor.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Q50
 
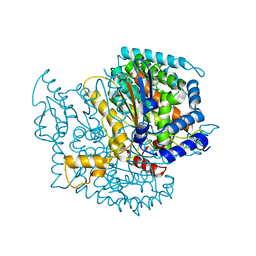 | |
1Q54
 
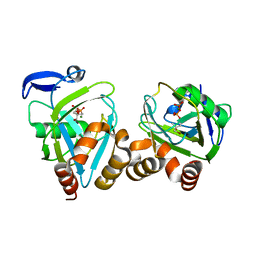 | | STRUCTURE AND MECHANISM OF ACTION OF ISOPENTENYLPYROPHOSPHATE-DIMETHYLALLYLPYROPHOSPHATE ISOMERASE: COMPLEX WITH THE BROMOHYDRINE OF IPP | | Descriptor: | 4-BROMO-3-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oudjama, Y, Ghosh, S, Stalon, V, Droogmans, L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure and mechanism of action of isopentenylpyrophosphate-dimethylallylpyrophosphate isomerase.
J.Am.Chem.Soc., 125, 2003
|
|
1Q57
 
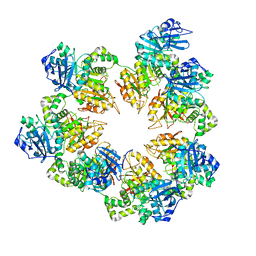 | | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7 | | Descriptor: | DNA primase/helicase | | Authors: | Toth, E.A, Li, Y, Sawaya, M.R, Cheng, Y, Ellenberger, T. | | Deposit date: | 2003-08-06 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The Crystal Structure of the Bifunctional Primase-Helicase of Bacteriophage T7
Mol.Cell, 12, 2003
|
|
1Q5D
 
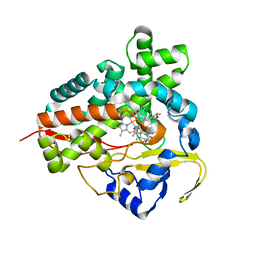 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1Q5E
 
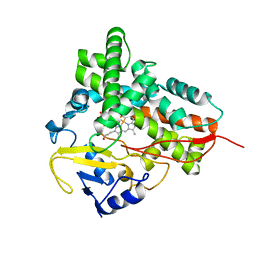 | | Substrate-free Cytochrome P450epoK | | Descriptor: | P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
1Q5H
 
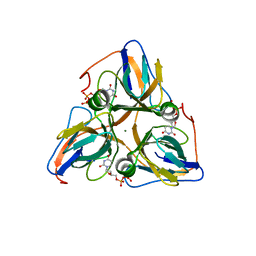 | | Human dUTP Pyrophosphatase complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, dUTP pyrophosphatase | | Authors: | Mol, C.D, Harris, J.M, McIntosh, E.M, Tainer, J.A. | | Deposit date: | 2003-08-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human dUTP pyrophosphatase: uracil recognition by a Beta hairpin and active sites formed by three separate subunits
Structure, 4, 1996
|
|
1Q5M
 
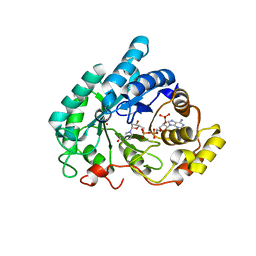 | | Binary complex of rabbit 20alpha-hydroxysteroid dehydrogenase with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin-E2 9-reductase, SULFATE ION | | Authors: | Couture, J.F, Legrand, P, Cantin, L, Labrie, F, Luu-The, V, Breton, R. | | Deposit date: | 2003-08-08 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Loop Relaxation, A Mechanism that Explains the Reduced Specificity of Rabbit 20alpha-Hydroxysteroid Dehydrogenase, A Member of the Aldo-Keto Reductase Superfamily.
J.Mol.Biol., 339, 2004
|
|
1Q5N
 
 | |
1Q5O
 
 | | HCN2J 443-645 in the presence of cAMP, selenomethionine derivative | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Zagotta, W.N, Olivier, N.B, Black, K.D, Young, E.C, Olson, R, Gouaux, J.E. | | Deposit date: | 2003-08-08 | | Release date: | 2003-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | STRUCTURAL BASIS FOR MODULATION AND AGONIST SPECIFICITY OF HCN PACEMAKER CHANNELS
Nature, 425, 2003
|
|
1Q5T
 
 | | Gln48 PLA2 separated from Vipoxin from the venom of Vipera ammodytes meridionalis. | | Descriptor: | Phospholipase A2 inhibitor, SULFATE ION | | Authors: | Georgieva, D.N, Perbandt, M, Rypniewski, W, Hristov, K, Genov, N, Betzel, C. | | Deposit date: | 2003-08-11 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray structure of a snake venom Gln48 phospholipase A2 at 1.9A resolution reveals
anion-binding sites.
Biochem.Biophys.Res.Commun., 316, 2004
|
|
1Q5U
 
 | | HUMAN DUTP PYROPHOSPHATASE | | Descriptor: | dUTP pyrophosphatase | | Authors: | Mol, C.D, Harris, J.M, Mcintosh, E.M, Tainer, J.A. | | Deposit date: | 2003-08-11 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human dUTP Pyrophosphatase: Uracil Recognition by a Beta Hairpin and Active Sites Formed by Three Separate Subunits
Structure, 4, 1996
|
|
1Q5V
 
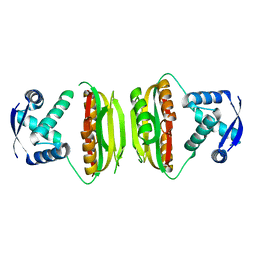 | | Apo-NikR | | Descriptor: | Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
1Q5X
 
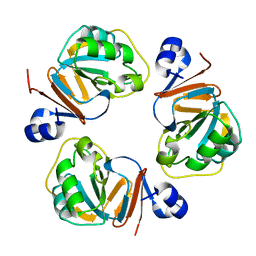 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
1Q5Y
 
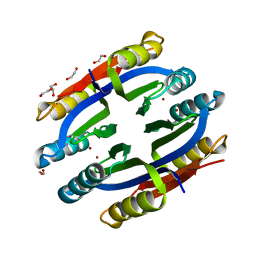 | | Nickel-Bound C-terminal Regulatory Domain of NikR | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Nickel responsive regulator | | Authors: | Schreiter, E.R, Sintchak, M.D, Guo, Y, Chivers, P.T, Sauer, R.T, Drennan, C.L. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the Nickel-Responsive Transcription Factor NikR
Nat.Struct.Biol., 10, 2003
|
|
1Q5Z
 
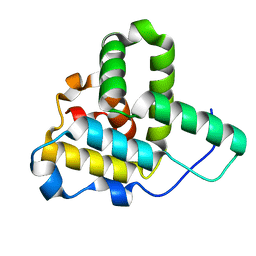 | | Crystal Structure of the C-terminal Actin Binding Domain of Salmonella Invasion Protein A (SipA) | | Descriptor: | SipA | | Authors: | Stebbins, C.E, Lilic, M, Galkin, V.E, Orlova, A, VanLoock, M.S, Egelman, E.H. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms.
Science, 301, 2003
|
|
1Q61
 
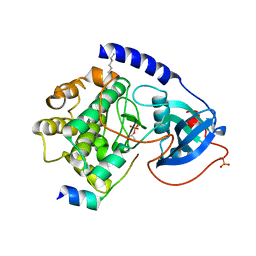 | | PKA triple mutant model of PKB | | Descriptor: | N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1Q62
 
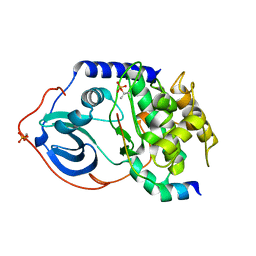 | | PKA double mutant model of PKB | | Descriptor: | cAMP-dependent protein kinase inhibitor, alpha form, cAMP-dependent protein kinase, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1Q63
 
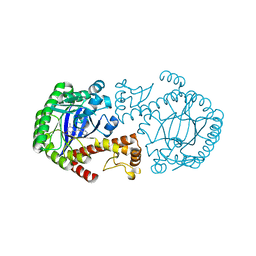 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-Diamino-8-(1H-imidazol-2-ylsulfanylmethyl)-3H-quinazoline-4-one crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(1H-IMIDAZOL-2-YLSULFANYLMETHYL)-3H-QUINAZOLINE-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1Q65
 
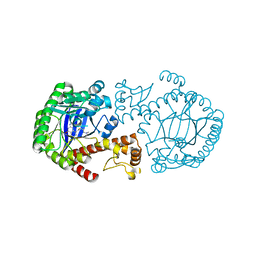 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2,6-DIAMINO-8-(2-dimethylaminoethylsulfanylmethyl)-3H-QUINAZOLIN-4-ONE crystallized at pH 5.5 | | Descriptor: | 2,6-DIAMINO-8-(2-DIMETHYLAMINOETHYLSULFANYLMETHYL)-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1Q66
 
 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-6-AMINOMETHYL-8-phenylsulfanylmethyl-3H-QUINAZOLIN-4-ONE crystallized at pH 5.5 | | Descriptor: | 2-AMINO-6-AMINOMETHYL-8-PHENYLSULFANYLMETHYL-3H-QUINAZOLIN-4-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Brenk, R, Meyer, E, Reuter, K, Stubbs, M.T, Garcia, G.A, Klebe, G. | | Deposit date: | 2003-08-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic Study of Inhibitors of tRNA-guanine Transglycosylase Suggests a New Structure-based Pharmacophore for Virtual Screening.
J.Mol.Biol., 338, 2004
|
|
1Q67
 
 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
1Q6C
 
 | | Crystal Structure of Soybean Beta-Amylase Complexed with Maltose | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-amylase | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6D
 
 | | Crystal structure of Soybean Beta-Amylase Mutant (M51T) with Increased pH Optimum | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
1Q6E
 
 | | Crystal Structure of Soybean Beta-Amylase Mutant (E178Y) with Increased pH Optimum at pH 5.4 | | Descriptor: | SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Hirata, A, Adachi, M, Sekine, A, Kang, Y.N, Utsumi, S, Mikami, B. | | Deposit date: | 2003-08-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Enzymatic Analysis of Soybean {beta}-Amylase Mutants with Increased pH Optimum
J.Biol.Chem., 279, 2004
|
|
