1PWL
 
 | | Crystal structure of human Aldose Reductase complexed with NADP and Minalrestat | | Descriptor: | 2[4-BROMO-2-FLUOROPHENYL)METHYL]-6-FLUOROSPIRO[ISOQUINOLINE-4-(1H),3'-PYRROLIDINE]-1,2',3,5'(2H)-TETRONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
1PWM
 
 | | Crystal structure of human Aldose Reductase complexed with NADP and Fidarestat | | Descriptor: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
1PWO
 
 | |
1PWP
 
 | | Crystal Structure of the Anthrax Lethal Factor complexed with Small Molecule Inhibitor NSC 12155 | | Descriptor: | Lethal factor, N,N'-BIS(4-AMINO-2-METHYLQUINOLIN-6-YL)UREA, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of small molecule inhibitors of anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PWQ
 
 | | Crystal structure of Anthrax Lethal Factor complexed with Thioacetyl-Tyr-Pro-Met-Amide, a metal-chelating peptidyl small molecule inhibitor | | Descriptor: | Lethal factor, N-(SULFANYLACETYL)TYROSYLPROLYLMETHIONINAMIDE, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PWU
 
 | | Crystal Structure of Anthrax Lethal Factor complexed with (3-(N-hydroxycarboxamido)-2-isobutylpropanoyl-Trp-methylamide), a known small molecule inhibitor of matrix metalloproteases. | | Descriptor: | 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, Lethal factor, ZINC ION | | Authors: | Wong, T.Y, Schwarzenbacher, R, Liddington, R.C. | | Deposit date: | 2003-07-02 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for substrate and inhibitor selectivity of the anthrax lethal factor.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1PWV
 
 | |
1PWW
 
 | |
1PWX
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with bromide | | Descriptor: | BROMIDE ION, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1PWY
 
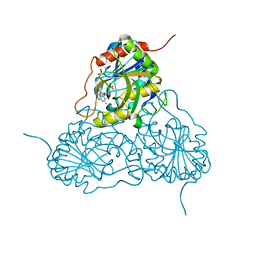 | | CRYSTAL STRUCTURE OF HUMAN PNP COMPLEXED WITH ACYCLOVIR | | Descriptor: | 9-HYROXYETHOXYMETHYLGUANINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Dos Santos, D.M, Canduri, F, Pereira, J.H, Vinicius Bertacine Dias, M, Silva, R.G, Mendes, M.A, Palma, M.S, Basso, L.A, De Azevedo, W.F, Santos, D.S. | | Deposit date: | 2003-07-02 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human purine nucleoside phosphorylase complexed with acyclovir.
Biochem.Biophys.Res.Commun., 308, 2003
|
|
1PWZ
 
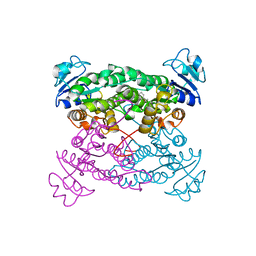 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with (R)-styrene oxide and chloride | | Descriptor: | CHLORIDE ION, R-STYRENE OXIDE, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1PX0
 
 | | Crystal structure of the haloalcohol dehalogenase HheC complexed with the haloalcohol mimic (R)-1-para-nitro-phenyl-2-azido-ethanol | | Descriptor: | (R)-1-PARA-NITRO-PHENYL-2-AZIDO-ETHANOL, halohydrin dehalogenase | | Authors: | de Jong, R.M, Tiesinga, J.J.W, Rozeboom, H.J, Kalk, K.H, Tang, L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2003-07-02 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of a Bacterial Haloalcohol Dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site
EMBO J., 22, 2003
|
|
1PX2
 
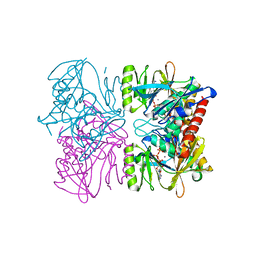 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP (Form 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Synapsin I | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-07-02 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
1PX3
 
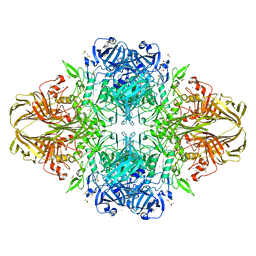 | | E. COLI (LACZ) BETA-GALACTOSIDASE (G794A) | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Juers, D.H, Hakda, S, Matthews, B.W, Huber, R.E. | | Deposit date: | 2003-07-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Altered Activity of Gly794 Variants of Escherichia coli Beta-Galactosidase
Biochemistry, 42, 2003
|
|
1PX4
 
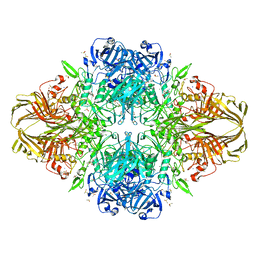 | | E. COLI (LACZ) BETA-GALACTOSIDASE (G794A) WITH IPTG BOUND | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Hakda, S, Matthews, B.W, Huber, R.E. | | Deposit date: | 2003-07-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Altered Activity of Gly794 Variants of Escherichia coli Beta-Galactosidase
Biochemistry, 42, 2003
|
|
1PX5
 
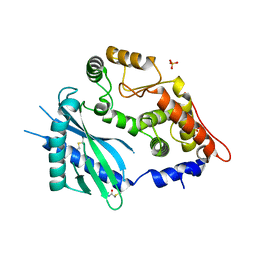 | | Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase | | Descriptor: | 2'-5'-oligoadenylate synthetase 1, SULFATE ION | | Authors: | Hartmann, R, Justesen, J, Sarkar, S.N, Sen, G.C, Yee, V.C. | | Deposit date: | 2003-07-02 | | Release date: | 2003-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of the 2'-specific and double-stranded RNA-activated interferon-induced antiviral protein 2'-5'-oligoadenylate synthetase
Mol.Cell, 12, 2003
|
|
1PX6
 
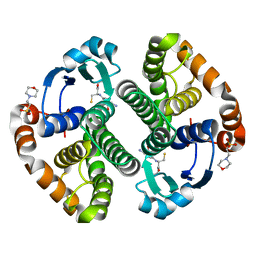 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to asparagine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase P | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1PX7
 
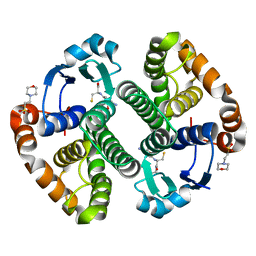 | | A folding mutant of human class pi glutathione transferase, created by mutating aspartate 153 of the wild-type protein to glutamate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUTATHIONE, ... | | Authors: | Kong, G.K.-W, Polekhina, G, McKinstry, W.J, Parker, M.W, Dragani, B, Aceto, A, Paludi, D, Principe, D.R, Mannervik, B, Stenberg, G. | | Deposit date: | 2003-07-02 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The multi-functional role of a highly conserved aspartic acid residue in glutathione transferase P1-1
To be Published
|
|
1PX8
 
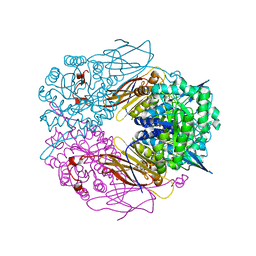 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | Beta-xylosidase, beta-D-xylopyranose | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-03 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1PXD
 
 | | Crystal structure of the complex of jacalin with meso-tetrasulphonatophenylporphyrin. | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, Agglutinin alpha chain, Agglutinin beta-3 chain | | Authors: | Goel, M, Anuradha, P, Kaur, K.J, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2003-07-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding to jacalin is facilitated by the inherent plasticity of the carbohydrate-binding site: novel mode of lectin-ligand interaction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PXG
 
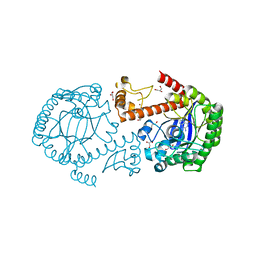 | | Crystal structure of the mutated tRNA-guanine transglycosylase (TGT) D280E complexed with preQ1 | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Kittendorf, J.D, Sgraja, T, Reuter, K, Klebe, G, Garcia, G.A. | | Deposit date: | 2003-07-04 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An essential role for aspartate 264 in catalysis by tRNA-guanine transglycosylase from Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
1PXI
 
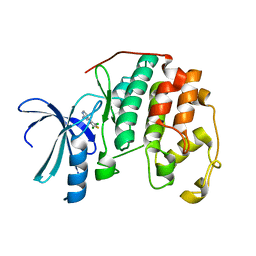 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,5-Dichloro-thiophen-3-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,5-DICHLOROTHIEN-3-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1PXJ
 
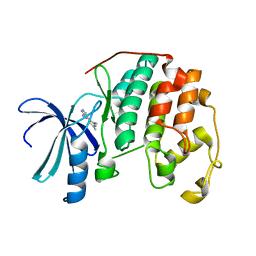 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR 4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-ylamine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1PXK
 
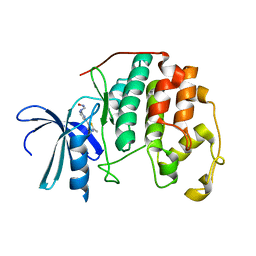 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR N-[4-(2,4-Dimethyl-thiazol-5-yl)pyrimidin-2-yl]-N'-hydroxyiminoformamide | | Descriptor: | Cell division protein kinase 2, N-[4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)PYRIMIDIN-2-YL]-N'-HYDROXYIMIDOFORMAMIDE | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
1PXL
 
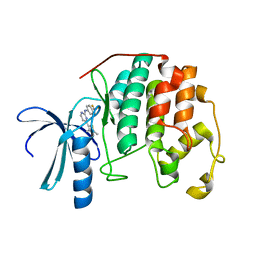 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR [4-(2,4-Dimethyl-thiazol-5-yl)-pyrimidin-2-yl]-(4-trifluoromethyl-phenyl)-amine | | Descriptor: | 4-(2,4-DIMETHYL-1,3-THIAZOL-5-YL)-N-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE, Cell division protein kinase 2 | | Authors: | Wu, S.Y, McNae, I, Kontopidis, G, McClue, S.J, McInnes, C, Stewart, K.J, Wang, S, Zheleva, D.I, Marriage, H, Lane, D.P, Taylor, P, Fischer, P.M, Walkinshaw, M.D. | | Deposit date: | 2003-07-04 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a novel family of CDK inhibitors with the program LIDAEUS: structural basis for ligand-induced disordering of the activation loop
Structure, 11, 2003
|
|
