1NEG
 
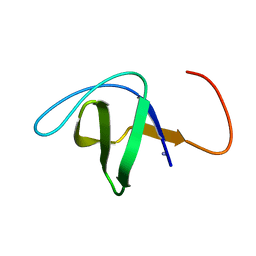 | | Crystal Structure Analysis of N-and C-terminal labeled SH3-domain of alpha-Chicken Spectrin | | Descriptor: | AZIDE ION, Spectrin alpha chain, brain | | Authors: | Mueller, U, Buessow, K, Diehl, A, Niesen, F.H, Nyarsik, L, Heinemann, U. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid purification and crystal structure analysis of a small protein carrying two terminal affinity tags
J.STRUCT.FUNCT.GENOM., 4, 2003
|
|
1NEJ
 
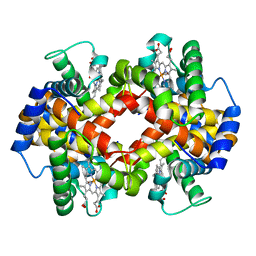 | | Crystalline Human Carbonmonoxy Hemoglobin S (Liganded Sickle Cell Hemoglobin) Exhibits The R2 Quaternary State At Neutral pH In The Presence Of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-12-11 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1NEP
 
 | | Crystal Structure Analysis of the Bovine NPC2 (Niemann-Pick C2) Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epididymal secretory protein E1, PHOSPHATE ION | | Authors: | Friedland, N, Liou, H.-L, Lobel, P, Stock, A.M. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Cholesterol-binding Protein Deficient in Niemann-Pick Type C2 Disease
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NES
 
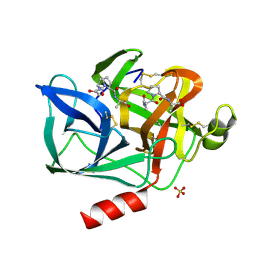 | | STRUCTURE OF THE PRODUCT COMPLEX OF ACETYL-ALA-PRO-ALA WITH PORCINE PANCREATIC ELASTASE AT 1.65 ANGSTROMS RESOLUTION | | Descriptor: | ACETYL-ALA-PRO-ALA, CALCIUM ION, ELASTASE, ... | | Authors: | Meyer Junior, E.F, Radhakrishnan, R, M Cole, G, Presta, L.G. | | Deposit date: | 1995-07-31 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the product complex of acetyl-Ala-Pro-Ala with porcine pancreatic elastase at 1.65 A resolution.
J.Mol.Biol., 189, 1986
|
|
1NEX
 
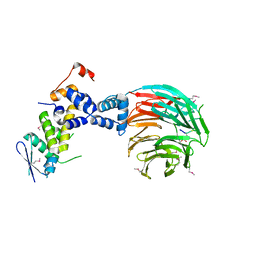 | | Crystal Structure of ScSkp1-ScCdc4-CPD peptide complex | | Descriptor: | CDC4 protein, Centromere DNA-binding protein complex CBF3 subunit D, GLL(TPO)PPQSG | | Authors: | Orlicky, S, Tang, X, Willems, A, Tyers, M, Sicheri, F. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Phosphodependent Substrate Selection and
Orientation by the SCFCdc4 Ubiquitin Ligase
Cell(Cambridge,Mass.), 112, 2003
|
|
1NEY
 
 | | Triosephosphate Isomerase in Complex with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | Authors: | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | Deposit date: | 2002-12-12 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NF0
 
 | | Triosephosphate Isomerase in Complex with DHAP | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, triosephosphate isomerase | | Authors: | Jogl, G, Rozovsky, S, McDermott, A.E, Tong, L. | | Deposit date: | 2002-12-12 | | Release date: | 2003-01-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimal alignment for enzymatic proton transfer: Structure of the
Michaelis complex of triosephosphate isomerase at 1.2-A resolution
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NF2
 
 | |
1NF3
 
 | | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6 | | Descriptor: | G25K GTP-binding protein, placental isoform, MAGNESIUM ION, ... | | Authors: | Garrard, S.M, Capaldo, C.T, Gao, L, Rosen, M.K, Macara, I.G, Tomchick, D.R. | | Deposit date: | 2002-12-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6
Embo J., 22, 2003
|
|
1NF4
 
 | | X-Ray Structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different states (reduced structure) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (II) ION, SULFATE ION, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NF5
 
 | | Crystal Structure of Lactose Synthase, Complex with Glucose | | Descriptor: | 1,2-ETHANEDIOL, Alpha-lactalbumin, CALCIUM ION, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2002-12-13 | | Release date: | 2002-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lactose Synthase Reveals a Large Conformational Change in its Catalytic Component, the beta-1,4-galactosyltransferase
J.Mol.Biol., 310, 2001
|
|
1NF6
 
 | | X-ray structure of the Desulfovibrio desulfuricans bacterioferritin: the diiron site in different catalytic states ("cycled" structure: reduced in solution and allowed to reoxidise before crystallisation) | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, FE (III) ION, GLYCEROL, ... | | Authors: | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | Deposit date: | 2002-12-13 | | Release date: | 2003-04-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
1NF7
 
 | |
1NF8
 
 | | Crystal structure of PhzD protein active site mutant with substrate | | Descriptor: | (5S,6S)-5-[(1-carboxyethenyl)oxy]-6-hydroxycyclohexa-1,3-diene-1-carboxylic acid, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1NF9
 
 | | Crystal Structure of PhzD protein from Pseudomonas aeruginosa | | Descriptor: | FORMIC ACID, octyl beta-D-glucopyranoside, phenazine biosynthesis protein phzD | | Authors: | Parsons, F, Calabrese, K, Eisenstein, E, Ladner, J.E. | | Deposit date: | 2002-12-13 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and mechanism of Pseudomonas aeruginosa PhzD, an isochorismatase from the phenazine biosynthetic pathway
Biochemistry, 42, 2003
|
|
1NFB
 
 | |
1NFF
 
 | | Crystal structure of Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-14 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFG
 
 | | Structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Xu, Z, Yang, Y, Jiang, W, Arnold, E, Ding, J. | | Deposit date: | 2002-12-14 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of D-Hydantoinase from Burkholderia pickettii at a Resolution of 2.7 Angstroms: Insights into the Molecular Basis of Enzyme Thermostability.
J.Bacteriol., 185, 2003
|
|
1NFH
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | Descriptor: | conserved hypothetical protein AF1956 | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2002-12-15 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
1NFJ
 
 | | Structure of a Sir2 substrate, alba, reveals a mechanism for deactylation-induced enhancement of DNA-binding | | Descriptor: | conserved hypothetical protein AF1956 | | Authors: | Zhao, K, Chai, X, Marmorstein, R. | | Deposit date: | 2002-12-15 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Sir2 substrate, alba, reveals a mechanism for deacetylation-induced enhancement of DNA-binding
J.Biol.Chem., 278, 2003
|
|
1NFN
 
 | | APOLIPOPROTEIN E3 (APOE3) | | Descriptor: | APOLIPOPROTEIN E3 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NFO
 
 | | APOLIPOPROTEIN E2 (APOE2, D154A MUTATION) | | Descriptor: | APOLIPOPROTEIN E2 | | Authors: | Rupp, B, Parkin, S. | | Deposit date: | 1996-07-17 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel mechanism for defective receptor binding of apolipoprotein E2 in type III hyperlipoproteinemia.
Nat.Struct.Biol., 3, 1996
|
|
1NFQ
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Androsterone, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-15 | | Release date: | 2002-12-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFR
 
 | | Rv2002 gene product from Mycobacterium tuberculosis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative oxidoreductase Rv2002 | | Authors: | Yang, J.K, Park, M.S, Waldo, G.S, Suh, S.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-12-16 | | Release date: | 2002-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Directed evolution approach to a structural genomics project: Rv2002 from Mycobacterium tuberculosis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NFS
 
 | |
