1M27
 
 | | Crystal structure of SAP/FynSH3/SLAM ternary complex | | Descriptor: | CITRATE ANION, Proto-oncogene tyrosine-protein kinase FYN, SH2 domain protein 1A, ... | | Authors: | Chan, B, Griesbach, J, Song, H.K, Poy, F, Terhorst, C, Eck, M.J. | | Deposit date: | 2002-06-21 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | SAP couples Fyn to SLAM immune receptors.
NAT.CELL BIOL., 5, 2003
|
|
1M2I
 
 | | Crystal structure of E44A/E56A mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, J, Wang, Y.-H, Gan, J.-H, Wang, W.-H, Sun, B.-Y, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-06-24 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Cytochrome b5 Mutated at the Charged Surface-Residues and Their Interactions with Cytochrome c
Chin.J.Chem., 20, 2002
|
|
1M2M
 
 | | Crystal structure of E44A/E48A/E56A/D60A mutant of cytochrome b5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome b5 | | Authors: | Wu, J, Wang, Y.-H, Gan, J.-H, Wang, W.-H, Sun, B.-Y, Huang, Z.-X, Xia, Z.-X. | | Deposit date: | 2002-06-24 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Cytochrome b5 Mutated at the Charged Surface-Residues and Their Interactions with Cytochrome c
Chin.J.Chem., 20, 2002
|
|
1M2T
 
 | | Mistletoe Lectin I from Viscum album in Complex with Adenine Monophosphate. Crystal Structure at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Krauspenhaar, R, Rypniewski, W, Kalkura, N, Moore, K, DeLucas, L, Stoeva, S, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 2002-06-25 | | Release date: | 2003-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystallisation under microgravity of mistletoe lectin I from Viscum album with adenine monophosphate and the crystal structure at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1M2W
 
 | | Pseudomonas fluorescens mannitol 2-dehydrogenase ternary complex with NAD and D-mannitol | | Descriptor: | D-MANNITOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, mannitol dehydrogenase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-06-25 | | Release date: | 2002-11-15 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Pseudomonas fluorescens Mannitol 2-Dehydrogenase Binary and Ternary Complexes. Specificity and Catalytic Mechanism
J.Biol.Chem., 277, 2002
|
|
1M2X
 
 | |
1M2Z
 
 | | Crystal structure of a dimer complex of the human glucocorticoid receptor ligand-binding domain bound to dexamethasone and a TIF2 coactivator motif | | Descriptor: | DEXAMETHASONE, glucocorticoid receptor, nuclear receptor coactivator 2, ... | | Authors: | Bledsoe, R.B, Montana, V.G, Stanley, T.B, Delves, C.J, Apolito, C.J, Mckee, D.D, Consler, T.G, Parks, D.J, Stewart, E.L, Willson, T.M, Lambert, M.H, Moore, J.T, Pearce, K.H, Xu, H.E. | | Deposit date: | 2002-06-26 | | Release date: | 2003-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Glucocorticoid Receptor Ligand Binding Domain Reveals a Novel Mode of Receptor Dimerization and Coactivator Recognition
Cell(Cambridge,Mass.), 110, 2002
|
|
1M32
 
 | | Crystal Structure of 2-aminoethylphosphonate Transaminase | | Descriptor: | 2-aminoethylphosphonate-pyruvate aminotransferase, PHOSPHATE ION, PHOSPHONOACETALDEHYDE, ... | | Authors: | Chen, C.C.H, Zhang, H, Kim, A.D, Howard, A, Sheldrick, G.M, Mariano-Dunnaway, D, Herzberg, O. | | Deposit date: | 2002-06-26 | | Release date: | 2002-11-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Degradation Pathway of the Phosphonate Ciliatine: Crystal Structure of 2-Aminoethylphosphonate Transaminase
Biochemistry, 41, 2002
|
|
1M33
 
 | | Crystal Structure of BioH at 1.7 A | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-PROPANOIC ACID, BioH protein | | Authors: | Sanishvili, R, Savchenko, A, Skarina, T, Edwards, A, Joachimiak, A, Yakunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-26 | | Release date: | 2003-01-21 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Integrating structure, bioinformatics, and enzymology to discover function: BioH, a new carboxylesterase from Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
1M35
 
 | | Aminopeptidase P from Escherichia coli | | Descriptor: | AMINOPEPTIDASE P, MANGANESE (II) ION | | Authors: | Graham, S.C, Lee, M, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-06-27 | | Release date: | 2003-05-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1M38
 
 | | Structure of Inorganic Pyrophosphatase | | Descriptor: | COBALT (II) ION, INORGANIC PYROPHOSPHATASE, PHOSPHATE ION | | Authors: | Kuranova, I.P, Polyakov, K.M, Levdikov, V.M, Smirnova, E.A, Hohne, W.E, Lamzin, V.S, Meijers, R. | | Deposit date: | 2002-06-27 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of Saccharomyces cerevisiae inorganic pyrophosphatase complexed with cobalt and phosphate ions
CRYSTALLOGRAPHY REPORTS, 48, 2003
|
|
1M3D
 
 | | Structure of Type IV Collagen NC1 Domains | | Descriptor: | BROMIDE ION, GLYCEROL, LUTETIUM (III) ION, ... | | Authors: | Sundaramoorthy, M, Meiyappan, M, Todd, P, Hudson, B.G. | | Deposit date: | 2002-06-27 | | Release date: | 2003-01-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of NC1 Domains. Structural Basis for Type IV Collagen Assembly in Basement Membranes
J.Biol.Chem., 277, 2002
|
|
1M3E
 
 | |
1M3H
 
 | | Crystal Structure of Hogg1 D268E Mutant with Product Oligonucleotide | | Descriptor: | 5'-D(P*GP*CP*GP*TP*CP*CP*AP*(DDX))-3', 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*TP*AP*CP*C)-3', ... | | Authors: | Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-06-27 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of End Products Resulting from Lesion Processing by a DNA Glycosylase/Lyase
Chem.Biol., 11, 2004
|
|
1M3K
 
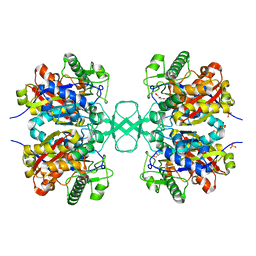 | | biosynthetic thiolase, inactive C89A mutant | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-06-28 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M3Q
 
 | |
1M3U
 
 | | Crystal Structure of Ketopantoate Hydroxymethyltransferase complexed the Product Ketopantoate | | Descriptor: | 3-methyl-2-oxobutanoate hydroxymethyltransferase, KETOPANTOATE, MAGNESIUM ION | | Authors: | von Delft, F, Inoue, T, Saldanha, S.A, Ottenhof, H.H, Dhanaraj, V, Witty, M, Abell, C, Smith, A.G, Blundell, T.L. | | Deposit date: | 2002-06-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E. coli Ketopantoate Hydroxymethyl Transferase Complexed with Ketopantoate and Mg(2+), Solved by Locating 160 Selenomethionine Sites.
Structure, 11, 2003
|
|
1M3W
 
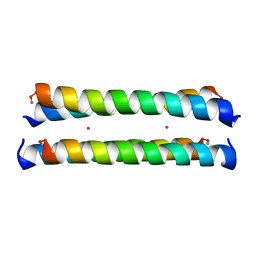 | | Crystal Structure of a Molecular Maquette Scaffold | | Descriptor: | H10H24, MERCURY (II) ION | | Authors: | Huang, S.S, Gibney, B.R, Stayrook, S.E, Dutton, P.L, Lewis, M. | | Deposit date: | 2002-07-01 | | Release date: | 2003-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Structure of a Maquette Scaffold
J.Mol.Biol., 326, 2003
|
|
1M3X
 
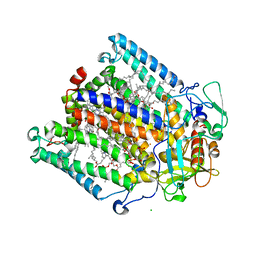 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Camara-Artigas, A, Brune, D, Allen, J.P. | | Deposit date: | 2002-07-01 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Interactions between lipids and bacterial reaction centers determined by protein crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1M3Z
 
 | | Biosynthetic thiolase, C89A mutant, complexed with acetyl coenzyme A | | Descriptor: | ACETYL COENZYME *A, Acetyl-CoA acetyltransferase, SULFATE ION | | Authors: | Kursula, P, Ojala, J, Lambeir, A.-M, Wierenga, R.K. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The catalytic cycle of biosynthetic thiolase: A conformational
journey of an acetyl group through four binding modes and two oxyanion holes
Biochemistry, 41, 2002
|
|
1M40
 
 | | ULTRA HIGH RESOLUTION CRYSTAL STRUCTURE OF TEM-1 | | Descriptor: | BETA-LACTAMASE TEM, PHOSPHATE ION, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE, ... | | Authors: | Minasov, G, Wang, X, Shoichet, B.K. | | Deposit date: | 2002-07-01 | | Release date: | 2002-07-17 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | An ultrahigh resolution structure of TEM-1 beta-lactamase suggests a role for Glu166 as the general base in acylation.
J.Am.Chem.Soc., 124, 2002
|
|
1M44
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-APO Structure | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
1M47
 
 | | Crystal Structure of Human Interleukin-2 | | Descriptor: | SULFATE ION, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, Wells, J.A, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Binding of small molecules to an adaptive protein-protein interface.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M48
 
 | | Crystal Structure of Human IL-2 Complexed with (R)-N-[2-[1-(Aminoiminomethyl)-3-piperidinyl]-1-oxoethyl]-4-(phenylethynyl)-L-phenylalanine methyl ester | | Descriptor: | 2-[3-METHYL-4-(N-METHYL-GUANIDINO)-BUTYRYLAMINO]-3-(4-PHENYLETHYNYL-PHENYL)-PROPIONIC ACID METHYL ESTER, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1M49
 
 | | Crystal Structure of Human Interleukin-2 Complexed with SP-1985 | | Descriptor: | 2-[2-(1-CARBAMIMIDOYL-PIPERIDIN-3-YL)-ACETYLAMINO]-3-{4-[2-(3-OXALYL-1H-INDOL-7-YL)ETHYL]-PHENYL}-PROPIONIC ACID METHYL ESTER, interleukin-2 | | Authors: | Arkin, M.A, Randal, M, DeLano, W.L, Hyde, J, Luong, T.N, Oslob, J.D, Raphael, D.R, Taylor, L, Wang, J, McDowell, R.S, Wells, J.A, Braisted, A.C. | | Deposit date: | 2002-07-02 | | Release date: | 2002-07-31 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of small molecules to an adaptive
protein-protein interface
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
