1KZE
 
 | | Complex of MBP-C and bivalent Man-terminated glycopeptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
1KZG
 
 | | DbsCdc42(Y889F) | | Descriptor: | CDC42 HOMOLOG, GUANINE NUCLEOTIDE EXCHANGE FACTOR DBS | | Authors: | Rossman, K.L, Worthylake, D.K, Snyder, J.T, Siderovski, D.P, Campbell, S.L, Sondek, J. | | Deposit date: | 2002-02-06 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange.
EMBO J., 21, 2002
|
|
1KZH
 
 | | Structure of a pyrophosphate-dependent phosphofructokinase from the Lyme disease spirochete Borrelia burgdorferi | | Descriptor: | SULFATE ION, phosphofructokinase | | Authors: | Moore, S.A, Ronimus, R.S, Roberson, R.S, Morgan, H.W. | | Deposit date: | 2002-02-06 | | Release date: | 2002-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of a pyrophosphate-dependent phosphofructokinase from the Lyme disease spirochete Borrelia burgdorferi.
Structure, 10, 2002
|
|
1KZI
 
 | | Crystal Structure of EcTS/dUMP/THF Complex | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (6S)-5,6,7,8-TETRAHYDROFOLATE, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, ... | | Authors: | Fritz, T.A, Liu, L, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access.
Biochemistry, 41, 2002
|
|
1KZJ
 
 | | Crystal Structure of EcTS W80G/dUMP/CB3717 Complex | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Fritz, T.A, Liu, L, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Tryptophan 80 and leucine 143 are critical for the hydride transfer step of thymidylate synthase by controlling active site access.
Biochemistry, 41, 2002
|
|
1KZK
 
 | | JE-2147-HIV Protease Complex | | Descriptor: | (4R)-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-N-(2-methylbenzyl)-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Reiling, K.K, Endres, N.F, Dauber, D.S, Craik, C.S, Stroud, R.M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Anisotropic Dynamics of the JE-2147-HIV Protease Complex:
Drug Resistance and Thermodynamic Binding Mode Examined in a 1.09 A Structure
Biochemistry, 41, 2002
|
|
1KZM
 
 | | Distal Heme Pocket Mutant (R38S/H42E) of Recombinant Horseradish Peroxidase C (HRP C). | | Descriptor: | CACODYLATE ION, CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Meno, K, Henriksen, A, Smith, A.T, Gajhede, M. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the two horseradish peroxidase catalytic residue variants H42E and R38S/H42E: implications for the catalytic cycle.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KZN
 
 | | Crystal Structure of E. coli 24kDa Domain in Complex with Clorobiocin | | Descriptor: | CLOROBIOCIN, DNA GYRASE SUBUNIT B | | Authors: | Lafitte, D, Lamour, V, Tsvetkov, P.O, Makarov, A.A, Klich, M, Deprez, P, Moras, D, Briand, C, Gilli, R. | | Deposit date: | 2002-02-07 | | Release date: | 2002-06-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5'-methyl group of the noviose.
Biochemistry, 41, 2002
|
|
1KZO
 
 | | PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH FARNESYLATED K-RAS4B PEPTIDE PRODUCT AND FARNESYL DIPHOSPHATE SUBSTRATE BOUND SIMULTANEOUSLY | | Descriptor: | ACETIC ACID, FARNESYL, FARNESYL DIPHOSPHATE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Reaction Path of Protein Farnesyltransferase at Atomic Resolution
Nature, 419, 2002
|
|
1KZP
 
 | | PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH A FARNESYLATED K-RAS4B PEPTIDE PRODUCT | | Descriptor: | ACETIC ACID, FARNESYL, Farnesylated K-Ras4B peptide product, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2002-02-07 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reaction path of protein farnesyltransferase at atomic resolution
Nature, 419, 2002
|
|
1KZQ
 
 | |
1KZU
 
 | | INTEGRAL MEMBRANE PERIPHERAL LIGHT HARVESTING COMPLEX FROM RHODOPSEUDOMONAS ACIDOPHILA STRAIN 10050 | | Descriptor: | BACTERIOCHLOROPHYLL A, LIGHT HARVESTING PROTEIN B-800/850, Rhodopin b-D-glucoside | | Authors: | Cogdell, R.J, Freer, A.A, Isaacs, N.W, Hawthornthwaite-Lawless, A.M, Mcdermott, G, Papiz, M.Z, Prince, S.M. | | Deposit date: | 1996-08-31 | | Release date: | 1997-04-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Apoprotein structure in the LH2 complex from Rhodopseudomonas acidophila strain 10050: modular assembly and protein pigment interactions.
J.Mol.Biol., 268, 1997
|
|
1L00
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
1L01
 
 | |
1L02
 
 | |
1L04
 
 | |
1L05
 
 | |
1L08
 
 | |
1L09
 
 | |
1L0A
 
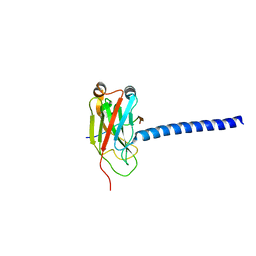 | | DOWNSTREAM REGULATOR TANK BINDS TO THE CD40 RECOGNITION SITE ON TRAF3 | | Descriptor: | TNF receptor associated factor 3, TRAF family member-associated NF-kappa-b activator | | Authors: | Li, C, Ni, C.-Z, Havert, M.L, Cabezas, E, He, J, Kaiser, D, Reed, J.C, Satterthwait, A.C, Cheng, G, Ely, K.R. | | Deposit date: | 2002-02-08 | | Release date: | 2002-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Downstream regulator TANK binds to the CD40 recognition site on TRAF3.
Structure, 10, 2002
|
|
1L0D
 
 | |
1L0E
 
 | |
1L0F
 
 | |
1L0G
 
 | |
1L0J
 
 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-11 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
