1J4R
 
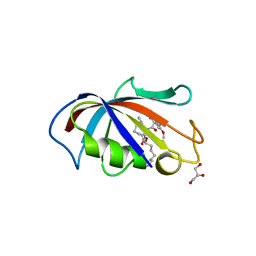 | | FK506 BINDING PROTEIN COMPLEXED WITH FKB-001 | | Descriptor: | 1-[2,2-DIFLUORO-2-(3,4,5-TRIMETHOXY-PHENYL)-ACETYL]-PIPERIDINE-2-CARBOXYLIC ACID 4-PHENYL-1-(3-PYRIDIN-3-YL-PROPYL)-BUTYL ESTER, FK506-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2001-10-29 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-Aryl-2,2-difluoroacetamide FKBP12 ligands: synthesis and X-ray structural studies.
Org.Lett., 3, 2001
|
|
1J4S
 
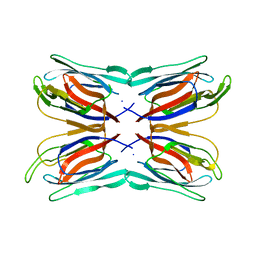 | | Structure of Artocarpin: a Lectin with Mannose Specificity (Form 1) | | Descriptor: | Artocarpin | | Authors: | Pratap, J.V, Jeyaprakash, A.A, Rani, P.G, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2001-10-30 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of artocarpin, a Moraceae lectin with mannose specificity, and its complex with methyl-alpha-D-mannose: implications to the generation of carbohydrate specificity.
J.Mol.Biol., 317, 2002
|
|
1J4T
 
 | | Structure of Artocarpin: a Lectin with Mannose Specificity (Form 2) | | Descriptor: | Artocarpin | | Authors: | Pratap, J.V, Jeyaprakash, A.A, Rani, P.G, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2001-10-30 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of artocarpin, a Moraceae lectin with mannose specificity, and its complex with methyl-alpha-D-mannose: implications to the generation of carbohydrate specificity.
J.Mol.Biol., 317, 2002
|
|
1J4U
 
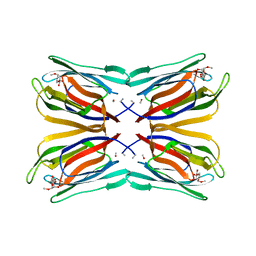 | | Structure of Artocarpin Complexed with Me-alpha-Mannose | | Descriptor: | Artocarpin, methyl alpha-D-mannopyranoside | | Authors: | Pratap, J.V, Jeyaprakash, A.A, Rani, P.G, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2001-10-30 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of artocarpin, a Moraceae lectin with mannose specificity, and its complex with methyl-alpha-D-mannose: implications to the generation of carbohydrate specificity.
J.Mol.Biol., 317, 2002
|
|
1J52
 
 | |
1J53
 
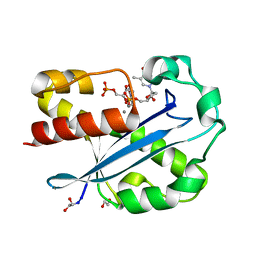 | | Structure of the N-terminal Exonuclease Domain of the Epsilon Subunit of E.coli DNA Polymerase III at pH 8.5 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1J54
 
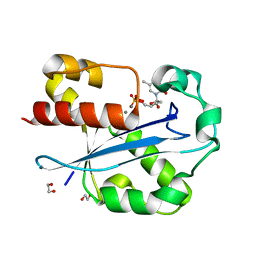 | | Structure of the N-terminal exonuclease domain of the epsilon subunit of E.coli DNA polymerase III at pH 5.8 | | Descriptor: | 1,2-ETHANEDIOL, DNA polymerase III, epsilon chain, ... | | Authors: | Hamdan, S, Carr, P.D, Brown, S.E, Ollis, D.L, Dixon, N.E. | | Deposit date: | 2002-01-22 | | Release date: | 2002-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Proofreading during Replication of the Escherichia coli Chromosome
Structure, 10, 2002
|
|
1J58
 
 | | Crystal Structure of Oxalate Decarboxylase | | Descriptor: | FORMIC ACID, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Anand, R, Dorrestein, P.C, Kinsland, C, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-02-25 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 A resolution.
Biochemistry, 41, 2002
|
|
1J59
 
 | | CATABOLITE GENE ACTIVATOR PROTEIN (CAP)/DNA COMPLEX + ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE | | Descriptor: | 5'-D(*AP*TP*AP*TP*GP*TP*CP*AP*CP*AP*CP*TP*TP*TP*TP*CP*G )-3', 5'-D(*GP*CP*GP*AP*AP*AP*AP*GP*TP*GP*TP*GP*AP*C)-3', ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Parkinson, G, Wilson, C, Gunasekera, A, Ebright, Y.W, Ebright, R.H, Berman, H.M. | | Deposit date: | 2002-03-01 | | Release date: | 2002-03-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface.
J.Mol.Biol., 260, 1996
|
|
1J5E
 
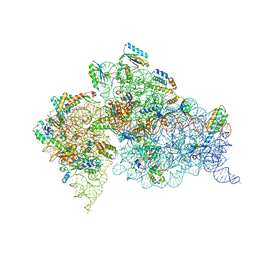 | | Structure of the Thermus thermophilus 30S Ribosomal Subunit | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Wimberly, B.T, Brodersen, D.E, Clemons Jr, W.M, Morgan-Warren, R, Carter, A.P, Vonrhein, C, Hartsch, T, Ramakrishnan, V. | | Deposit date: | 2002-04-08 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the 30S ribosomal subunit.
Nature, 407, 2000
|
|
1J5O
 
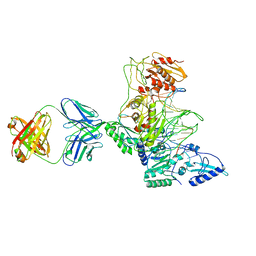 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1J5P
 
 | |
1J5S
 
 | |
1J5T
 
 | |
1J5U
 
 | |
1J5W
 
 | |
1J5X
 
 | |
1J5Y
 
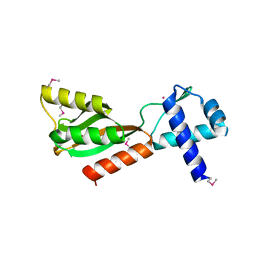 | |
1J6O
 
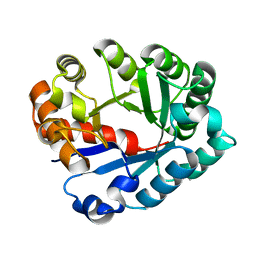 | |
1J6P
 
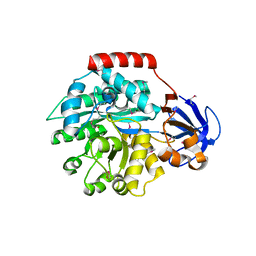 | |
1J6R
 
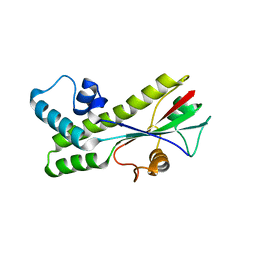 | |
1J6S
 
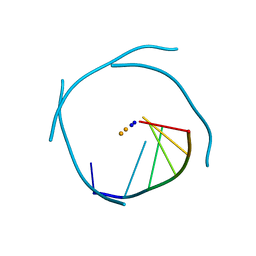 | | Crystal Structure of an RNA Tetraplex (UGAGGU)4 with A-tetrads, G-tetrads, U-tetrads and G-U octads | | Descriptor: | 5'-R(*(BRUP*GP*AP*GP*GP*U)-3', BARIUM ION, SODIUM ION | | Authors: | Pan, B, Xiong, Y, Shi, K, Deng, J, Sundaralingam, M. | | Deposit date: | 2002-07-10 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA purine-rich tetraplex containing adenine tetrads:
implications for specific binding in RNA tetraplexes
Structure, 11, 2003
|
|
1J6U
 
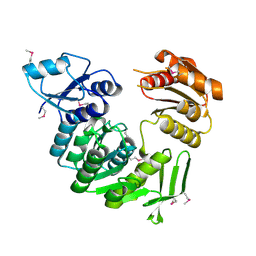 | |
1J6V
 
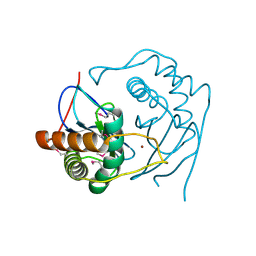 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, C2 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
1J6W
 
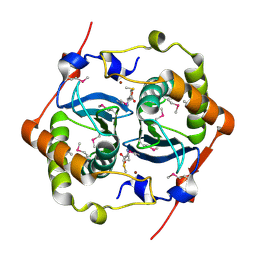 | | CRYSTAL STRUCTURE OF HAEMOPHILUS INFLUENZAE LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
