1HP5
 
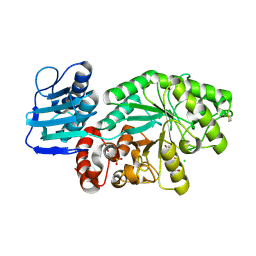 | | STREPTOMYCES PLICATUS BETA-N-ACETYLHEXOSAMINIDASE COMPLEXED WITH INTERMEDIATE ANALOUGE NAG-THIAZOLINE | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, BETA-N-ACETYLHEXOSAMINIDASE, CHLORIDE ION, ... | | Authors: | Mark, B.L. | | Deposit date: | 2000-12-12 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis in a bacterial beta-hexosaminidase.
J.Biol.Chem., 276, 2001
|
|
1HP7
 
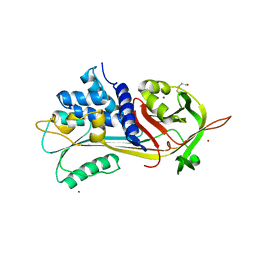 | | A 2.1 ANGSTROM STRUCTURE OF AN UNCLEAVED ALPHA-1-ANTITRYPSIN SHOWS VARIABILITY OF THE REACTIVE CENTER AND OTHER LOOPS | | Descriptor: | ALPHA-1-ANTITRYPSIN, BETA-MERCAPTOETHANOL, ZINC ION | | Authors: | Kim, S.-J, Woo, J.-R, Seo, E.J, Yu, M.-H, Ryu, S.-E. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-14 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A 2.1 A resolution structure of an uncleaved alpha(1)-antitrypsin shows variability of the reactive center and other loops.
J.Mol.Biol., 306, 2001
|
|
1HPL
 
 | |
1HPT
 
 | |
1HPU
 
 | | 5'-NUCLEOTIDASE (CLOSED FORM), COMPLEX WITH AMPCP | | Descriptor: | 5'-NUCLEOTIDASE, MANGANESE (II) ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Knoefel, T, Straeter, N. | | Deposit date: | 2000-12-13 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of hydrolysis of phosphate esters by the dimetal center of 5'-nucleotidase based on crystal structures.
J.Mol.Biol., 309, 2001
|
|
1HPZ
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
1HQ0
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF E.COLI CYTOTOXIC NECROTIZING FACTOR TYPE 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2000-12-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
1HQ1
 
 | | STRUCTURAL AND ENERGETIC ANALYSIS OF RNA RECOGNITION BY A UNIVERSALLY CONSERVED PROTEIN FROM THE SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Sagar, M.B, Doudna, J.A. | | Deposit date: | 2000-12-13 | | Release date: | 2001-01-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle.
J.Mol.Biol., 307, 2001
|
|
1HQ2
 
 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX OF E.COLI HPPK(R82A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.25 ANGSTROM RESOLUTION | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, ACETATE ION, ... | | Authors: | Blaszczyk, J, Ji, X. | | Deposit date: | 2000-12-13 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | Descriptor: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | Authors: | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | Deposit date: | 2000-12-14 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1HQ6
 
 | | STRUCTURE OF PYRUVOYL-DEPENDENT HISTIDINE DECARBOXYLASE AT PH 8 | | Descriptor: | HISTIDINE DECARBOXYLASE | | Authors: | Schelp, E, Worley, S, Monzingo, A.F, Ernst, S, Robertus, J.D. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | pH-induced structural changes regulate histidine decarboxylase activity in Lactobacillus 30a.
J.Mol.Biol., 306, 2001
|
|
1HQ7
 
 | |
1HQ8
 
 | | CRYSTAL STRUCTURE OF THE MURINE NK CELL-ACTIVATING RECEPTOR NKG2D AT 1.95 A | | Descriptor: | NKG2-D | | Authors: | Wolan, D.W, Teyton, L, Rudolph, M.G, Villmow, B, Bauer, S, Busch, D.H, Wilson, I.A. | | Deposit date: | 2000-12-14 | | Release date: | 2001-03-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A.
Nat.Immunol., 2, 2001
|
|
1HQC
 
 | | STRUCTURE OF RUVB FROM THERMUS THERMOPHILUS HB8 | | Descriptor: | ADENINE, MAGNESIUM ION, RUVB | | Authors: | Yamada, K, Kunishima, N, Mayanagi, K, Iwasaki, H, Morikawa, K. | | Deposit date: | 2000-12-15 | | Release date: | 2001-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HQD
 
 | | PSEUDOMONAS CEPACIA LIPASE COMPLEXED WITH TRANSITION STATE ANALOGUE OF 1-PHENOXY-2-ACETOXY BUTANE | | Descriptor: | (RP,SP)-O-(2R)-(1-PHENOXYBUT-2-YL)-METHYLPHOSPHONIC ACID CHLORIDE, CALCIUM ION, LIPASE | | Authors: | Luic, M, Tomic, S, Lescic, I, Ljubovic, E, Sepac, D, Sunjic, V, Vitale, L, Saenger, W, Kojic-Prodic, B. | | Deposit date: | 2000-12-15 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex of Burkholderia cepacia lipase with transition state analogue of 1-phenoxy-2-acetoxybutane: biocatalytic, structural and modelling study.
Eur.J.Biochem., 268, 2001
|
|
1HQE
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-15 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
1HQK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF LUMAZINE SYNTHASE FROM AQUIFEX AEOLICUS | | Descriptor: | 6,7-DIMETHYL-8-RIBITYLLUMAZINE SYNTHASE | | Authors: | Zhang, X, Meining, W, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 A resolution: determinants of thermostability revealed from structural comparisons.
J.Mol.Biol., 306, 2001
|
|
1HQL
 
 | | The xenograft antigen in complex with the B4 isolectin of Griffonia simplicifolia lectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LECTIN, ... | | Authors: | Tempel, W, Lipscomb, L.A, Rose, J.P, Woods, R.J. | | Deposit date: | 2000-12-18 | | Release date: | 2002-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The xenograft antigen bound to Griffonia simplicifolia lectin 1-B(4). X-ray crystal structure of the complex and molecular dynamics characterization of the binding site.
J.Biol.Chem., 277, 2002
|
|
1HQN
 
 | | THE SELENOMETHIONINE DERIVATIVE OF P3, THE MAJOR COAT PROTEIN OF THE LIPID-CONTAINING BACTERIOPHAGE PRD1. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | Benson, S.D, Bamford, J.K.H, Bamford, D.H, Burnett, R.M. | | Deposit date: | 2000-12-18 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1HQO
 
 | | CRYSTAL STRUCTURE OF THE NITROGEN REGULATION FRAGMENT OF THE YEAST PRION PROTEIN URE2P | | Descriptor: | URE2 PROTEIN | | Authors: | Umland, T.C, Taylor, K.L, Rhee, S, Wickner, R.B, Davies, D.R. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-14 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1HQQ
 
 | | MINIPROTEIN MP-2 (M9A) COMPLEX WITH STREPTAVIDIN | | Descriptor: | MP-2, STREPTAVIDIN | | Authors: | Yang, H.W, Liu, D.Q, Fan, X, White, M.A, Fox, R.O. | | Deposit date: | 2000-12-19 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conformational Ensemble Analysis of Ligand Binding in Streptavidin Mini-protein Complexes
To be Published
|
|
1HQR
 
 | | CRYSTAL STRUCTURE OF A SUPERANTIGEN BOUND TO THE HIGH-AFFINITY, ZINC-DEPENDENT SITE ON MHC CLASS II | | Descriptor: | HLA-DR ALPHA CHAIN, HLA-DR BETA CHAIN, MYELIN BASIC PROTEIN, ... | | Authors: | Li, Y, Li, H, Dimasi, N, Schlievert, P, Mariuzza, R. | | Deposit date: | 2000-12-19 | | Release date: | 2001-01-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a superantigen bound to the high-affinity, zinc-dependent site on MHC class II.
Immunity, 14, 2001
|
|
1HQS
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE DEHYDROGENASE FROM BACILLUS SUBTILIS | | Descriptor: | CITRIC ACID, ISOCITRATE DEHYDROGENASE, R-1,2-PROPANEDIOL, ... | | Authors: | Singh, S.K, Matsuno, K, LaPorte, D.C, Banaszak, L.J. | | Deposit date: | 2000-12-19 | | Release date: | 2001-07-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of Bacillus subtilis isocitrate dehydrogenase at 1.55 A. Insights into the nature of substrate specificity exhibited by Escherichia coli isocitrate dehydrogenase kinase/phosphatase.
J.Biol.Chem., 276, 2001
|
|
1HQT
 
 | | THE CRYSTAL STRUCTURE OF AN ALDEHYDE REDUCTASE Y50F MUTANT-NADP COMPLEX AND ITS IMPLICATIONS FOR SUBSTRATE BINDING | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Hyndman, D, Green, N.C, Li, L, Korithoski, B, Jia, Z, Flynn, T.G. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of an Aldehyde Reductase Y50F Mutant-NADP Complex and its Implications for Substrate Binding
Chem.Biol.Interact., 132, 2001
|
|
1HQU
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, POL POLYPROTEIN | | Authors: | Hsiou, Y, Ding, J, Arnold, E. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
