1FNF
 
 | |
1FNJ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88S/R90K | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1FNK
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88K/R90S | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1FNM
 
 | | STRUCTURE OF THERMUS THERMOPHILUS EF-G H573A | | Descriptor: | ELONGATION FACTOR G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Laurberg, M, Kristensen, O, Martemyanov, K, Gudkov, A.T, Nagaev, I, Hughes, D, Liljas, A. | | Deposit date: | 2000-08-22 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a mutant EF-G reveals domain III and possibly the fusidic acid binding site.
J.Mol.Biol., 303, 2000
|
|
1FNN
 
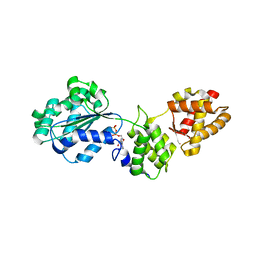 | | CRYSTAL STRUCTURE OF CDC6P FROM PYROBACULUM AEROPHILUM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION CONTROL PROTEIN 6, MAGNESIUM ION | | Authors: | Liu, J, Smith, C.L, DeRyckere, D, DeAngelis, K, Martin, G.S, Berger, J.M. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of Cdc6/Cdc18: implications for origin recognition and checkpoint control.
Mol.Cell, 6, 2000
|
|
1FNO
 
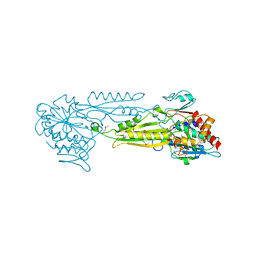 | | PEPTIDASE T (TRIPEPTIDASE) | | Descriptor: | PEPTIDASE T, SULFATE ION, ZINC ION | | Authors: | Hakansson, K, Miller, C.G. | | Deposit date: | 2000-08-22 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Peptidase T from Salmonella typhimurium
Eur.J.Biochem., 269, 2002
|
|
1FNT
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEASOME ACTIVATOR PROTEIN PA26, PROTEASOME COMPONENT C1, ... | | Authors: | Whitby, F.G, Masters, E, Kramer, L, Knowlton, J.R, Yao, Y, Wang, C.C, Hill, C.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activation of 20S proteasomes by 11S regulators.
Nature, 408, 2000
|
|
1FNY
 
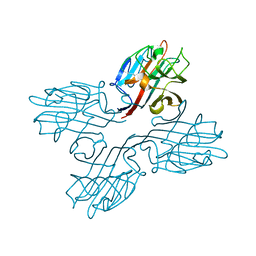 | | LEGUME LECTIN OF THE BARK OF ROBINIA PSEUDOACACIA. | | Descriptor: | BARK AGGLUTININ I,POLYPEPTIDE A, CALCIUM ION | | Authors: | Rabijns, A, Verboven, C, Rouge, P, Barre, A, Van Damme, E.J, Peumans, W.J, De Ranter, C.J. | | Deposit date: | 2000-08-24 | | Release date: | 2001-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of a legume lectin from the bark of Robinia pseudoacacia and its complex with N-acetylgalactosamine.
Proteins, 44, 2001
|
|
1FNZ
 
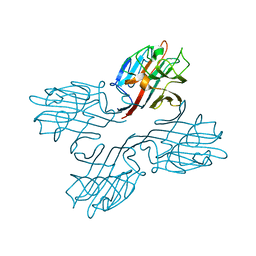 | | A bark lectin from robinia pseudoacacia in complex with N-acetylgalactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, BARK AGGLUTININ I, POLYPEPTIDE A, ... | | Authors: | Rabijns, A, Verboven, C, Rouge, P, Barre, A, Van Damme, E.J, Peumans, W.J, De Ranter, C.J. | | Deposit date: | 2000-08-24 | | Release date: | 2001-08-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a legume lectin from the bark of Robinia pseudoacacia and its complex with N-acetylgalactosamine
Proteins, 44, 2001
|
|
1FO0
 
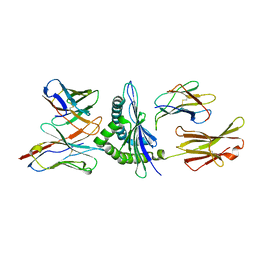 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | NATURALLY PROCESSED OCTAPEPTIDE PBM1, PROTEIN (ALLOGENEIC H-2KB MHC CLASS I MOLECULE), PROTEIN (BETA-2 MICROGLOBULIN), ... | | Authors: | Reiser, J.B, Darnault, C, Guimezanes, A, Gregoire, C, Mosser, T, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Malissen, B, Housset, D, Mazza, G. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a T cell receptor bound to an allogeneic MHC molecule.
Nat.Immunol., 1, 2000
|
|
1FO1
 
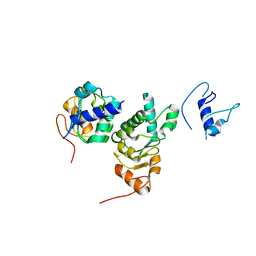 | |
1FO2
 
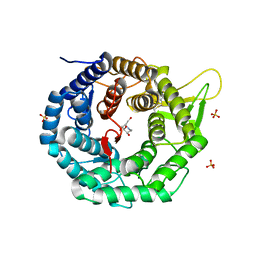 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE IN COMPLEX WITH 1-DEOXYMANNOJIRIMYCIN | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, ALPHA1,2-MANNOSIDASE, CALCIUM ION, ... | | Authors: | Vallee, F, Karaveg, K, Moremen, K.W, Herscovics, A, Howell, P.L. | | Deposit date: | 2000-08-24 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
1FO8
 
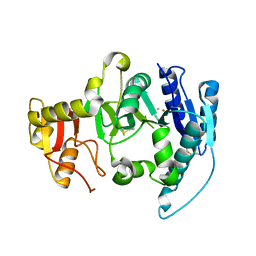 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, METHYL MERCURY ION | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FO9
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FOA
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FOB
 
 | |
1FOC
 
 | | Cytochrome C557: improperly folded thermus thermophilus C552 | | Descriptor: | CYTOCHROME RC557, HEME C | | Authors: | McRee, D.E, Williams, P.A, Fee, J.A, Bren, K.L. | | Deposit date: | 2000-08-27 | | Release date: | 2000-11-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recombinant cytochrome rC557 obtained from Escherichia coli cells expressing a truncated Thermus thermophilus cycA gene. Heme inversion in an improperly matured protein
J.Biol.Chem., 276, 2001
|
|
1FOI
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 1400W(H4B-FREE) | | Descriptor: | ACETATE ION, CACODYLATE ION, GLYCEROL, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors.
Biochemistry, 40, 2001
|
|
1FOJ
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 7-NITROINDAZOLE-2-CARBOXAMIDINE (H4B PRESENT) | | Descriptor: | 7-NITROINDAZOLE, 7-NITROINDAZOLE-2-CARBOXAMIDINE, ACETATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-11-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism
Biochemistry, 40, 2001
|
|
1FOL
 
 | | REDUCED BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH L-ARG(H4B-FREE) | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors
Biochemistry, 40, 2001
|
|
1FOO
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH L-ARG AND NO(H4B-FREE) | | Descriptor: | ACETATE ION, ARGININE, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors.
Biochemistry, 40, 2001
|
|
1FOP
 
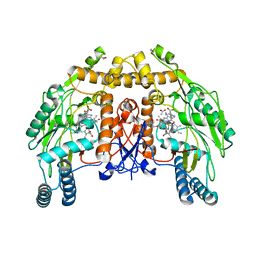 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH L-ARG AND NO(H4B-BOUND) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ARGININE, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 2000-08-28 | | Release date: | 2001-07-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic studies on endothelial nitric oxide synthase complexed with nitric oxide and mechanism-based inhibitors.
Biochemistry, 40, 2001
|
|
1FOT
 
 | |
1FOU
 
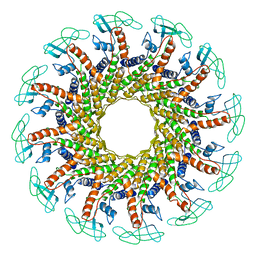 | | CONNECTOR PROTEIN FROM BACTERIOPHAGE PHI29 | | Descriptor: | UPPER COLLAR PROTEIN | | Authors: | Simpson, A.A, Tao, Y, Leiman, P.G, Badasso, M.O, He, Y, Jardine, P.J, Olson, N.H, Morais, M.C, Grimes, S.N, Anderson, D.L, Baker, T.S, Rossmann, M.G. | | Deposit date: | 2000-08-28 | | Release date: | 2000-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the bacteriophage phi29 DNA packaging motor.
Nature, 408, 2000
|
|
1FP1
 
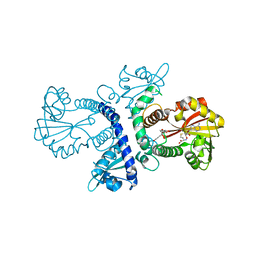 | | CRYSTAL STRUCTURE ANALYSIS OF CHALCONE O-METHYLTRANSFERASE | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, ISOLIQUIRITIGENIN 2'-O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2000-08-29 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases.
Nat.Struct.Biol., 8, 2001
|
|
