6WPB
 
 | |
6WPD
 
 | |
6WPO
 
 | |
6WPV
 
 | |
6WQE
 
 | | Solution Structure of the IWP-051-bound H-NOX from Shewanella woodyi in the Fe(II)CO ligation state | | Descriptor: | 5-fluoro-2-{1-[(2-fluorophenyl)methyl]-5-(1,2-oxazol-3-yl)-1H-pyrazol-3-yl}pyrimidin-4-ol, CARBON MONOXIDE, Heme NO binding domain protein, ... | | Authors: | Chen, C.Y, Lee, W, Montfort, W.R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051.
Protein Sci., 30, 2021
|
|
6WQJ
 
 | |
6WQL
 
 | |
6WQR
 
 | |
6WUX
 
 | |
6X1K
 
 | | Solution NMR structure of de novo designed TMB2.3 | | Descriptor: | De novo designed transmembrane beta-barrel TMB2.3 | | Authors: | Liang, B, Vorobieva, A.A, Chow, C.M, Baker, D, Tamm, L.K. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of transmembrane beta barrels.
Science, 371, 2021
|
|
6X46
 
 | | NMR solution structure of Asterix/Gtsf1 from mouse (CHHC zinc finger domains) | | Descriptor: | Gametocyte-specific factor 1, ZINC ION | | Authors: | Ipsaro, J.J, O'Brien, P.A, Bhattacharya, S, Palmer III, A.G, Joshua-Tor, L. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Asterix/Gtsf1 links tRNAs and piRNA silencing of retrotransposons.
Cell Rep, 34, 2021
|
|
6X4X
 
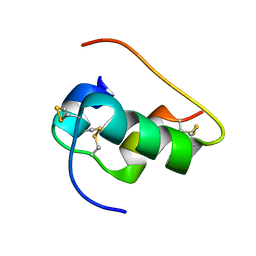 | | B24Y DKP insulin | | Descriptor: | Insulin, Insulin chain A | | Authors: | Weiss, M.A, Yang, Y. | | Deposit date: | 2020-05-24 | | Release date: | 2020-08-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Evolution of insulin at the edge of foldability and its medical implications.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6X63
 
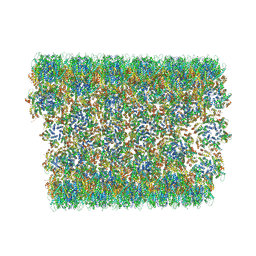 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6X6N
 
 | |
6X7I
 
 | |
6X8R
 
 | |
6XAH
 
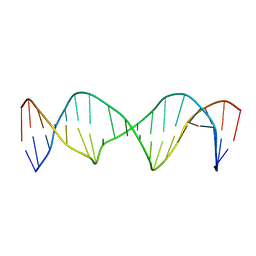 | | Structure of a Stable Interstrand DNA Crosslink Involving an dA Amino Group and an Abasic Site | | Descriptor: | DNA (5'-D(*TP*AP*GP*AP*TP*GP*AP*AP*CP*(AAB)P*TP*AP*GP*AP*CP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*GP*TP*CP*TP*AP*AP*GP*TP*TP*CP*AP*TP*CP*TP*A)-3') | | Authors: | Kellum Jr, A.H, Qiu, D, Voehler, M.W, Martin, W.J, Gates, K.S, Stone, M.P. | | Deposit date: | 2020-06-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable Interstrand DNA Cross-Link Involving a beta- N -Glycosyl Linkage Between an N 6 -dA Amino Group and an Abasic Site.
Biochemistry, 60, 2021
|
|
6XB7
 
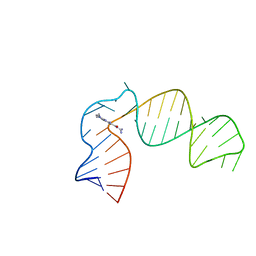 | | IRES-targeting Small Molecule Inhibits Enterovirus 71 Replication via Allosteric Stabilization of a Ternary Complex | | Descriptor: | 3-amino-N-(diaminomethylidene)-5-(dimethylamino)-6-(phenylethynyl)pyrazine-2-carboxamide, RNA (41-MER) | | Authors: | Tolbert, B.S, Davila, J, Sugarman, A.L, Chiu, L.Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex.
Nat Commun, 11, 2020
|
|
6XCR
 
 | | NMR structure of Ost4 in DPC micelles | | Descriptor: | Oligosaccharyltransferase | | Authors: | Chaudhary, B.P. | | Deposit date: | 2020-06-09 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR and MD simulations reveal the impact of the V23D mutation on the function of yeast oligosaccharyltransferase subunit Ost4.
Glycobiology, 31, 2021
|
|
6XCU
 
 | |
6XEH
 
 | |
6XFL
 
 | | Structural characterization of the type III secretion system pilotin-secretin complex InvH-InvG by NMR spectroscopy | | Descriptor: | Type 3 secretion system pilotin, Type 3 secretion system secretin | | Authors: | Majewski, D.D, Okon, M, Heinkel, F, Robb, C.S, Vuckovic, M, McIntosh, L.P, Strynadka, N.C.J. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Pilotin-Secretin Complex from the Salmonella enterica Type III Secretion System Using Hybrid Structural Methods.
Structure, 29, 2021
|
|
6XMN
 
 | |
6XN9
 
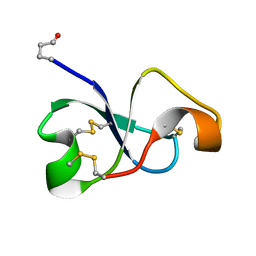 | | Solution NMR structure of recifin, a cysteine-rich tyrosyl-DNA Phosphodiesterase I modulatory peptide from the marine sponge Axinella sp. | | Descriptor: | Recifin modulatory peptide | | Authors: | Schroeder, C.I, Rosengren, K.J, O'Keefe, B.R. | | Deposit date: | 2020-07-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Recifin A, Initial Example of the Tyr-Lock Peptide Structural Family, Is a Selective Allosteric Inhibitor of Tyrosyl-DNA Phosphodiesterase I.
J.Am.Chem.Soc., 142, 2020
|
|
6XOR
 
 | |
