6V4I
 
 | | DanD | | Descriptor: | DanD peptide | | Authors: | Hoang, H.N. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | DanD
To Be Published
|
|
6V4T
 
 | | MPER-TMD of HIV-1 Env bound with the entry inhibitor S2C3 | | Descriptor: | 4,4'-(decane-1,10-diyl)bis(9-amino-2,3-dihydro-1H-cyclopenta[b]quinolin-4-ium), Envelope glycoprotein gp160 | | Authors: | Xiao, T, Frey, G, Fu, Q, Lavine, C.L, Scott, D.A, Seaman, M.S, Chou, J.J, Chen, B. | | Deposit date: | 2019-11-30 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HIV-1 fusion inhibitors targeting the membrane-proximal external region of Env spikes.
Nat.Chem.Biol., 16, 2020
|
|
6V5D
 
 | | EROS3 RDC and NOE Derived Ubiquitin Ensemble | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Smith, C.A, Griesinger, C, de Groot, B.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enhancing NMR derived ensembles with kinetics on multiple timescales.
J.Biomol.Nmr, 74, 2020
|
|
6V5L
 
 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
6V6T
 
 | |
6V88
 
 | |
6VA1
 
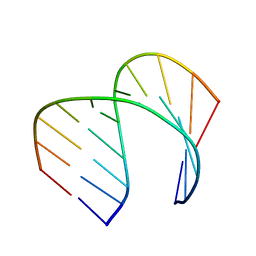 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element | | Descriptor: | RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA2
 
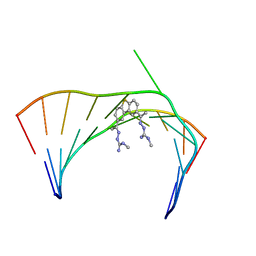 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MH5 | | Descriptor: | (2E,2'E)-2,2'-{dibenzo[b,d]thiene-2,8-diyldi[(1E)eth-1-yl-1-ylidene]}bis(N-methylhydrazine-1-carboximidamide), RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA3
 
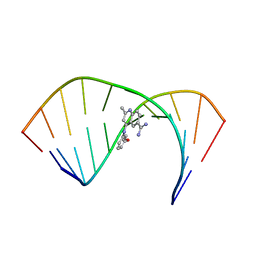 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MQC | | Descriptor: | 4-[(3-methoxyphenyl)amino]-2-methylquinoline-6-carboximidamide, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VA4
 
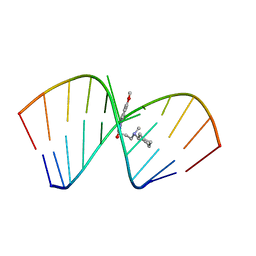 | | Solution Structure of the Tau pre-mRNA Exon 10 Splicing Regulatory Element Bound to MIP | | Descriptor: | N-[3-(8-methoxy-4-oxo-4,5-dihydro-3H-pyrimido[5,4-b]indol-3-yl)propyl]-N-methylcyclohexanaminium, RNA (5'-R(*CP*AP*CP*AP*CP*GP*UP*CP*GP*G)-3'), RNA (5'-R(*CP*CP*GP*GP*CP*AP*GP*UP*GP*UP*G)-3') | | Authors: | Chen, J.L, Fountain, M.A, Disney, M.D. | | Deposit date: | 2019-12-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Optimization, and Study of Small Molecules That Target Tau Pre-mRNA and Affect Splicing.
J.Am.Chem.Soc., 142, 2020
|
|
6VAR
 
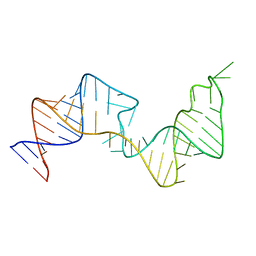 | | 61 nt human Hepatitis B virus epsilon pre-genomic RNA | | Descriptor: | RNA (61-MER) | | Authors: | LeBlanc, R.M, Kasprzak, W.K, Longhini, A.P, Abulwerdi, F, Ginocchio, S, Shields, B, Nyman, J, Svirydava, M, Del Vecchio, C, Ivanic, J, Schneekloth, J.S, Dayie, T.K, Shapiro, B.A, Le Grice, S.F.J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structural insights of the conserved "priming loop" of hepatitis B virus pre-genomic RNA.
J.Biomol.Struct.Dyn., 2021
|
|
6VE9
 
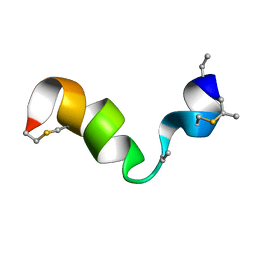 | |
6VED
 
 | | Solution structure of the TTD and linker region of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1 | | Authors: | Lemak, A, Houliston, S, Duan, S, Ong, M.S, Arrowsmith, C.H. | | Deposit date: | 2019-12-31 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
6VEE
 
 | |
6VFO
 
 | | Solution structure of the PHD of mouse UHRF1 (NP95) | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Lemak, A, Houliston, S, Duan, S, Arrowsmith, C.H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
6VG7
 
 | |
6VGA
 
 | |
6VGB
 
 | |
6VGT
 
 | |
6VH8
 
 | |
6VHJ
 
 | |
6VJQ
 
 | |
6VK2
 
 | |
6VL2
 
 | | Stigmurin | | Descriptor: | Stigmurin | | Authors: | Rodrigues, S.C.S, Resende, J.M, Araujo, R.M, Pedrosa, M.F.F. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR three-dimensional structure of the cationic peptide Stigmurin from Tityus stigmurus scorpion venom: In vitro antioxidant and in vivo antibacterial and healing activity.
Peptides, 137, 2021
|
|
6VLA
 
 | | Hs05 - Intragenic antimicrobial peptide | | Descriptor: | Intragenic antimicrobial peptide | | Authors: | Santos, M.A, Silva, E.M.C, Brand, G.D, Oliveira, A.L. | | Deposit date: | 2020-01-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of novel human Intragenic Antimicrobial Peptides, incorporation and release studies from ureasil-polyether hybrid matrix
Materials Science and Engineering C, Materials for Biological Applications, 119, 2020
|
|
