6GZK
 
 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, TMR3 (48-MER) | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
6GZN
 
 | |
6GZR
 
 | | Solution NMR structure of the tetramethylrhodamine (TMR) aptamer 3 in complex with 5-TAMRA | | Descriptor: | 5-carboxy methylrhodamine, tetramethylrhodamine aptamer | | Authors: | Duchardt-Ferner, E, Ohlenschlager, O, Kreutz, C.R, Wohnert, J. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine.
Nucleic Acids Res., 48, 2020
|
|
6H0I
 
 | | Solution structure of Melampsora larici-populina MlpP4.1 | | Descriptor: | Secreted protein | | Authors: | Tsan, P, Petre, B, Hecker, A, Rouhier, N, Duplessis, S. | | Deposit date: | 2018-07-09 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
6H0J
 
 | | A1-type ACP domain from module 5 of MLSA1 | | Descriptor: | Type I modular polyketide synthase | | Authors: | Moretto, L, Heylen, R, Holroyd, N, Vance, S, Broadhurst, R.W. | | Deposit date: | 2018-07-09 | | Release date: | 2019-03-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Modular type I polyketide synthase acyl carrier protein domains share a common N-terminally extended fold.
Sci Rep, 9, 2019
|
|
6H0Q
 
 | | B1-type ACP domain from module 7 of MLSB | | Descriptor: | Type I modular polyketide synthase | | Authors: | Moretto, L, Heylen, R, Holroyd, N, Vance, S, Broadhurst, R.W. | | Deposit date: | 2018-07-10 | | Release date: | 2019-03-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Modular type I polyketide synthase acyl carrier protein domains share a common N-terminally extended fold.
Sci Rep, 9, 2019
|
|
6H1K
 
 | | The major G-quadruplex form of HIV-1 LTR | | Descriptor: | DNA (28-MER) | | Authors: | Phan, A.T, Heddi, B, Butovskaya, E, Bakalar, B, Richter, S.N. | | Deposit date: | 2018-07-11 | | Release date: | 2018-10-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Major G-Quadruplex Form of HIV-1 LTR Reveals a (3 + 1) Folding Topology Containing a Stem-Loop.
J. Am. Chem. Soc., 140, 2018
|
|
6H3E
 
 | | Receptor-bound Ghrelin conformation | | Descriptor: | Appetite-regulating hormone, octan-1-amine | | Authors: | Ferre, G, Damian, M, M'Kadmi, C, Saurel, O, Czaplicki, G, Demange, P, Marie, J, Fehrentz, J.A, Baneres, J.L, Milon, A. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of G protein-coupled receptor-bound ghrelin reveal the critical role of the octanoyl chain.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6H5H
 
 | |
6H7I
 
 | |
6H7Q
 
 | |
6H8C
 
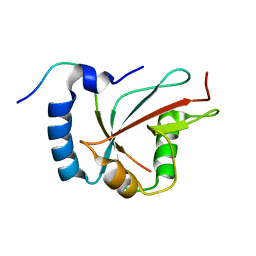 | | Structure of the human GABARAPL2 protein in complex with the UBA5 LIR motif | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 2, Ubiquitin-like modifier-activating enzyme 5 | | Authors: | Huber, J, Loehr, F, Gruber, J, Akutsu, M, Guentert, P, Doetsch, V, Rogov, V.V. | | Deposit date: | 2018-08-02 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An atypical LIR motif within UBA5 (ubiquitin like modifier activating enzyme 5) interacts with GABARAP proteins and mediates membrane localization of UBA5.
Autophagy, 16, 2020
|
|
6HAG
 
 | |
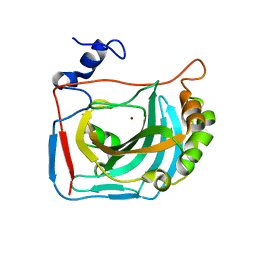
6HD2
 
 | |
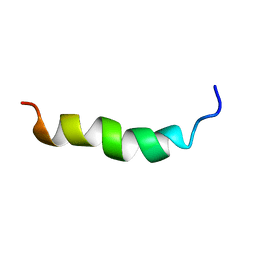
6HH0
 
 | |
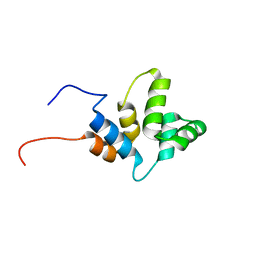
6HJ7
 
 | |
6HKA
 
 | |
6HKC
 
 | |
6HMI
 
 | |
6HMO
 
 | |
6HN9
 
 | | Nicomicin-1 -- Novel antimicrobial peptides from the Arctic polychaeta Nicomache minor provide new molecular insight into biological role of the BRICHOS domain | | Descriptor: | Nicomicin-1 | | Authors: | Panteleev, P.V, Tsarev, A.V, Bolosov, I.A, Paramonov, A.S, Marggraf, M.B, Sychev, S.V, Shenkarev, Z.O, Ovchinnikova, T.V. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Novel Antimicrobial Peptides from the Arctic PolychaetaNicomache minorProvide New Molecular Insight into Biological Role of the BRICHOS Domain.
Mar Drugs, 16, 2018
|
|
6HNE
 
 | |
6HNF
 
 | | Structure in solution of human fibronectin type III-domain 14 | | Descriptor: | Fibronectin | | Authors: | Zhong, X, Arnolds, O, Krenczyk, O, Gajewski, J, Puetz, S, Herrmann, C, Stoll, R. | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure in Solution of Fibronectin Type III Domain 14 Reveals Its Synergistic Heparin Binding Site.
Biochemistry, 57, 2018
|
|
6HNG
 
 | |
6HNH
 
 | |
