8JB4
 
 | | lipopolysaccharide-binding domain-LBDB | | Descriptor: | Antilipopolysaccharide factor D | | Authors: | Huang, J, Qin, Z. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-13 | | Last modified: | 2025-03-19 | | Method: | SOLUTION NMR | | Cite: | Machine learning and genetic algorithm-guided directed evolution for the development of antimicrobial peptides.
J Adv Res, 68, 2025
|
|
8JB9
 
 | | Solution structure of Anti-CRISPR protein AcrIIC5 | | Descriptor: | Type II-C anti-CRISPR protein, AcrIIC5 | | Authors: | Hong, S.H, Park, C, An, S.Y, Suh, J.Y. | | Deposit date: | 2023-05-08 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-21 | | Method: | SOLUTION NMR | | Cite: | Structural variants of AcrIIC5 inhibit Cas9 via divergent binding interfaces.
Structure, 33, 2025
|
|
8JFQ
 
 | | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-back Loop | | Descriptor: | 26mer-DNA | | Authors: | Liu, Y, Li, J, Zhang, Y, Wang, Y, Chen, J, Bian, Y, Xia, Y, Yang, M.H, Zheng, K, Wang, K.B, Kong, L.Y. | | Deposit date: | 2023-05-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major G-Quadruplex in the Human EGFR Oncogene Promoter Adopts a Unique Folding Topology with a Distinctive Snap-Back Loop.
J.Am.Chem.Soc., 145, 2023
|
|
8OFC
 
 | | Structure of an i-motif domain with the cytosine analog 1,3-diaza-2-oxophenoxacione (tC) at neutral pH | | Descriptor: | DNA (5'-D(*CP*(YCO)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*(DNR)P*GP*T)-3') | | Authors: | Mir, B, Serrano-Chacon, I, Terrazas, M, Gandioso, A, Garavis, M, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2023-03-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Site-specific incorporation of a fluorescent nucleobase analog enhances i-motif stability and allows monitoring of i-motif folding inside cells.
Nucleic Acids Res., 52, 2024
|
|
8OH7
 
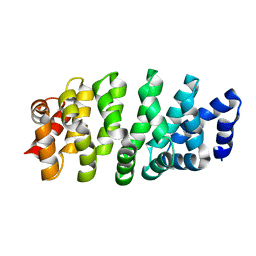 | |
8OIL
 
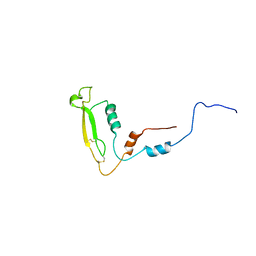 | |
8OJR
 
 | | Solution NMR Structure of Alpha-Synuclein 1-25 Peptide in 50% TFE. | | Descriptor: | Alpha-synuclein | | Authors: | Allen, S.G, Williams, C, Meade, R.M, Tang, T.M.S, Crump, M.P, Mason, J.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | An N-terminal alpha-Synuclein fragment binds lipid vesicles to modulate lipid induced aggregation
Cell Rep Phys Sci, 2023
|
|
8OL8
 
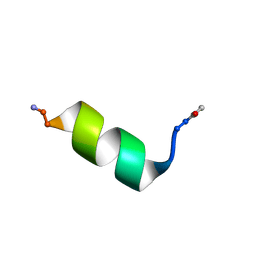 | |
8ONG
 
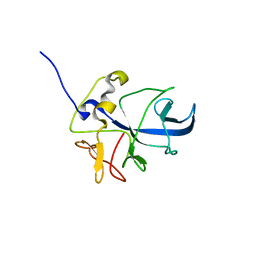 | |
8ONU
 
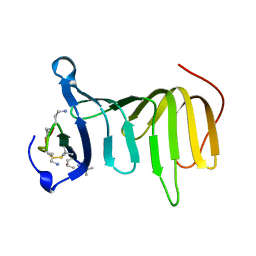 | |
8OR5
 
 | |
8OR8
 
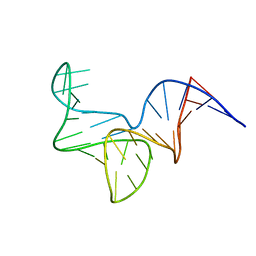 | |
8ORY
 
 | |
8ORZ
 
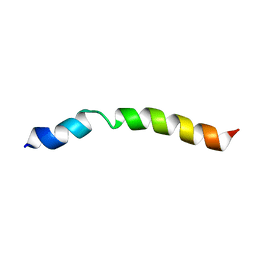 | |
8OS0
 
 | |
8OSQ
 
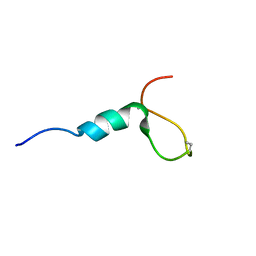 | |
8OVL
 
 | |
8OYD
 
 | |
8OZZ
 
 | |
8P25
 
 | | Solution structure of a chimeric U2AF2 RRM2 / FUBP1 N-Box | | Descriptor: | Splicing factor U2AF 65 kDa subunit,Far upstream element-binding protein 1 | | Authors: | Hipp, C, Sattler, M. | | Deposit date: | 2023-05-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | FUBP1 is a general splicing factor facilitating 3' splice site recognition and splicing of long introns.
Mol.Cell, 83, 2023
|
|
8P3A
 
 | | bacteriophage T5 l-alanoyl-d-glutamate peptidase Zn2+/Ca2+ form | | Descriptor: | CALCIUM ION, L-alanyl-D-glutamate peptidase, ZINC ION | | Authors: | Prokhorov, D.A, Kutyshenko, V.P, Mikoulinskaia, G.V. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-01-15 | | Method: | SOLUTION NMR | | Cite: | Conservative Tryptophan Residue in the Vicinity of an Active Site of the M15 Family l,d-Peptidases: A Key Element in the Catalysis.
Int J Mol Sci, 24, 2023
|
|
8POI
 
 | |
8S9E
 
 | | Solution structure of jarastatin (rJast), a disintegrin from Bothrops jararaca | | Descriptor: | Disintegrin jarastatin | | Authors: | Vasconcelos, A.A, Estrada, J.E.C, Zingali, R.B, Almeida, F.C.L. | | Deposit date: | 2023-03-28 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Toward the mechanism of jarastatin (rJast) inhibition of the integrin alpha V beta 3.
Int.J.Biol.Macromol., 255, 2024
|
|
8S9Y
 
 | | Taipan Natriuretic Peptide C -TNPc | | Descriptor: | Peptide TNP-c | | Authors: | Torres, A.M, Alewood, P.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Taipan Natriuretic Peptides Are Potent and Selective Agonists for the Natriuretic Peptide Receptor A.
Molecules, 28, 2023
|
|
8SCF
 
 | | TCEIII NMR Structure | | Descriptor: | RNA (30-MER) | | Authors: | Warden, M.S, Mueller, G.A, Hall, T.M.T. | | Deposit date: | 2023-04-05 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The translational repressor Glorund uses interchangeable RNA recognition domains to recognize Drosophila nanos.
Nucleic Acids Res., 51, 2023
|
|
