7TUJ
 
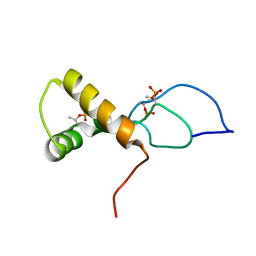 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
7TV5
 
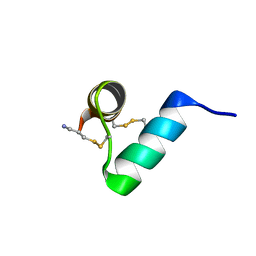 | | Disulfide-rich venom peptide lasiocepsin: P20A mutant | | Descriptor: | Lasiocepsin | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV6
 
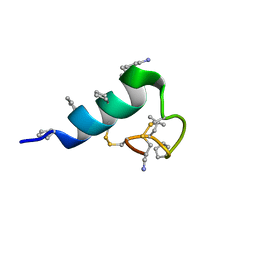 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: native loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV7
 
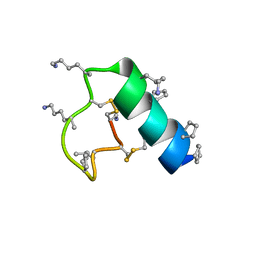 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: beta-3-Lys modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV8
 
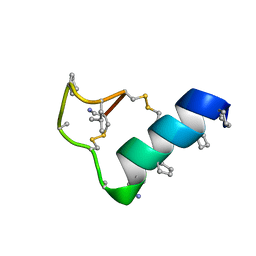 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: D-Ala modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TVQ
 
 | |
7TVR
 
 | |
7TXX
 
 | |
7TZ3
 
 | |
7TZ8
 
 | |
7TZD
 
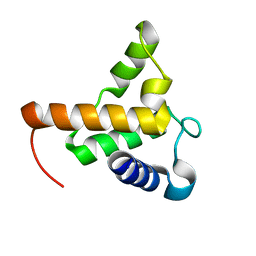 | |
7U37
 
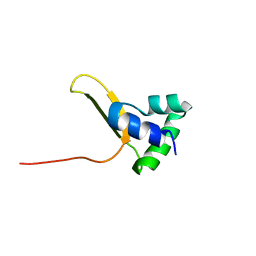 | |
7U8K
 
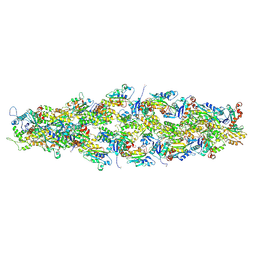 | | Magic Angle Spinning NMR Structure of Human Cofilin-2 Assembled on Actin Filaments | | Descriptor: | Actin, alpha skeletal muscle, Cofilin-2 | | Authors: | Kraus, J, Russell, R, Kudryashova, E, Xu, C, Katyal, N, Kudryashov, D, Perilla, J.R, Polenova, T. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | SOLID-STATE NMR | | Cite: | Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode.
Nat Commun, 13, 2022
|
|
7UBC
 
 | |
7UBD
 
 | |
7UBE
 
 | |
7UBF
 
 | |
7UBG
 
 | |
7UBH
 
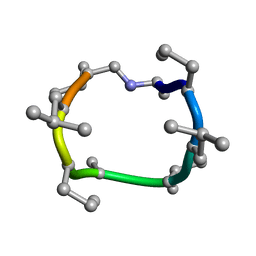 | |
7UBI
 
 | |
7UGA
 
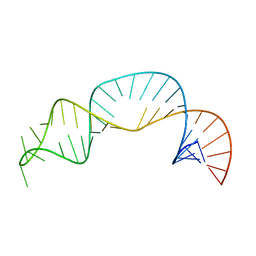 | | Solution structure of NPSL2 | | Descriptor: | NPSL2 RNA (43-MER) | | Authors: | Liu, Y, Keane, S.C. | | Deposit date: | 2022-03-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of NPSL2, A Regulatory Element in the oncomiR-1 RNA.
J.Mol.Biol., 434, 2022
|
|
7UGC
 
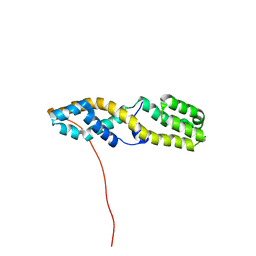 | | Structure of the N-terminal domain of ViaA | | Descriptor: | Protein ViaA | | Authors: | Lemak, A, Reichheld, S, Bhandari, V, Houliston, S, Arrowsmith, C.H, Sharpe, S, Houry, W.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of ViaA
To Be Published
|
|
7UH9
 
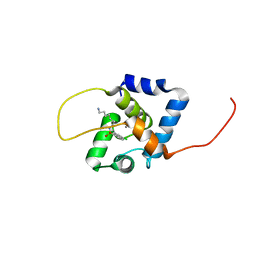 | | NMR structure of the cNTnC-cTnI chimera bound to W8 | | Descriptor: | CALCIUM ION, N-(7-aminoheptyl)-5-chloronaphthalene-1-sulfonamide, Troponin C, ... | | Authors: | Cai, F, Kampourakis, T, Cockburn, K.T, Sykes, B.D. | | Deposit date: | 2022-03-26 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Drugging the Sarcomere, a Delicate Balance: Position of N-Terminal Charge of the Inhibitor W7.
Acs Chem.Biol., 17, 2022
|
|
7UHA
 
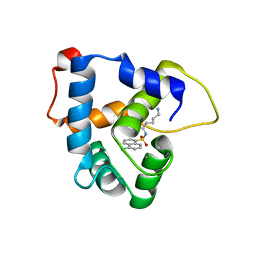 | | NMR structure of the cNTnC-cTnI chimera bound to W6 | | Descriptor: | CALCIUM ION, N-(5-aminopentyl)-5-chloronaphthalene-1-sulfonamide, Troponin C, ... | | Authors: | Cai, F, Kampourakis, T, Cockburn, K.T, Sykes, B.D. | | Deposit date: | 2022-03-26 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Drugging the Sarcomere, a Delicate Balance: Position of N-Terminal Charge of the Inhibitor W7.
Acs Chem.Biol., 17, 2022
|
|
7UI5
 
 | | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9 | | Descriptor: | CALCIUM ION, Protein S100-A9 | | Authors: | Reardon, P.N, Harman, J.L, Costello, S.M, Warren, G.D, Phillips, S.R, Connor, P.J, Marqusee, S, Harms, M.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
