7SUR
 
 | |
7SVC
 
 | | NMR structure of cTnC-TnI chimera bound to calcium and A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, CALCIUM ION, Troponin C, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-18 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SWG
 
 | | cTnC-TnI chimera complexed with A1 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3S)-7-fluoro-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SWI
 
 | | cTnC-TnI chimera complexed with A2 | | Descriptor: | 4-(3-cyano-3-methylazetidine-1-carbonyl)-N-[(3R,4S)-7-fluoro-4-hydroxy-6-methyl-3,4-dihydro-2H-1-benzopyran-3-yl]-5-methyl-1H-pyrrole-2-sulfonamide, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SWJ
 
 | | KirBac1.1 mutant - I131C | | Descriptor: | Inward rectifier potassium channel | | Authors: | Amani, R, Wylie, B.J. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Water Accessibility Refinement of the Extended Structure of KirBac1.1 in the Closed State.
Front Mol Biosci, 8, 2021
|
|
7SXB
 
 | |
7SXC
 
 | | cTnC-TnI chimera complexed with calcium | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles,Troponin I, ... | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SXD
 
 | | NMR solution structure TnC-TnI chimera | | Descriptor: | Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle chimera | | Authors: | Poppe, L, Hartman, J.J, Romero, A, Reagan, J.D. | | Deposit date: | 2021-11-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Thermodynamic Model for the Activation of Cardiac Troponin.
Biochemistry, 61, 2022
|
|
7SXI
 
 | | Solution Structure of Sds3 Capped Tudor Domain | | Descriptor: | Sin3 histone deacetylase corepressor complex component SDS3 | | Authors: | Marcum, R.D, Radhakrishnan, I. | | Deposit date: | 2021-11-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A capped Tudor domain within a core subunit of the Sin3L/Rpd3L histone deacetylase complex binds to nucleic acid G-quadruplexes.
J.Biol.Chem., 298, 2022
|
|
7T03
 
 | | NMR structure of a designed cold unfolding four helix bundle | | Descriptor: | Cold unfolding four helix bundle | | Authors: | Pulavarti, S, Szyperski, T, Yuen, S, Maguire, J, Griffin, J, Kuhlman, B. | | Deposit date: | 2021-11-29 | | Release date: | 2022-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | From Protein Design to the Energy Landscape of a Cold Unfolding Protein.
J.Phys.Chem.B, 126, 2022
|
|
7T1N
 
 | |
7T1O
 
 | |
7T1P
 
 | |
7T2F
 
 | | Solution structure of the model HEEH mini protein homodimer HEEH_TK_rd5_0341 | | Descriptor: | HEEH mini protein HEEH_TK_rd5_0341 | | Authors: | Lemak, A, Houliston, S, Kim, T.-E, Martel, C, Rocklin, G.J, Arrowsmith, C.H. | | Deposit date: | 2021-12-04 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dissecting the stability determinants of a challenging de novo protein fold using massively parallel design and experimentation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7T6G
 
 | | Truncated Ac-AIP-2 | | Descriptor: | Truncated Ac-AIP-2 | | Authors: | Daly, N.L, Cobos, C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-10-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Peptides derived from hookworm anti-inflammatory proteins suppress inducible colitis in mice and inflammatory cytokine production by human cells.
Front Med (Lausanne), 9, 2022
|
|
7T7W
 
 | |
7TA8
 
 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
7TB9
 
 | | Structural characterization of the biological synthetic peptide pCEMP1 | | Descriptor: | CEMP1-p1 | | Authors: | Lopez Giraldo, A, del Rio Portilla, F, Nidome Campos, M, Romo Arevalo, E, Arzate, H. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of cementum protein 1 derived peptide (CEMP1-p1) and its role in the mineralization process.
J.Pept.Sci., 29, 2023
|
|
7TH8
 
 | | Chickpea (Cicer arientinum) nodule-specific cysteine-rich peptide NCR13: Solution NMR structure of the isomer with C4:C10, C15:C30, and C23:C28 disulfide bonds | | Descriptor: | Nodule cysteine-rich protein 13 | | Authors: | Buchko, G.W, Zhou, M, Shah, D.M, Velivelli, S.L.S. | | Deposit date: | 2022-01-10 | | Release date: | 2022-02-02 | | Last modified: | 2025-01-08 | | Method: | SOLUTION NMR | | Cite: | Chickpea NCR13 disulfide cross-linking variants exhibit profound differences in antifungal activity and modes of action.
Plos Pathog., 20, 2024
|
|
7TIO
 
 | |
7TIP
 
 | |
7TIQ
 
 | |
7TIR
 
 | |
7TIS
 
 | |
7TOD
 
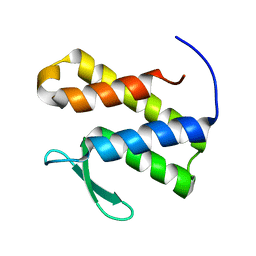 | | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA | | Descriptor: | Subversion of eukaryotic traffic protein A | | Authors: | Beck, W.H.J, Enoki, T.A, Feigenson, G.W, Nicholson, L, Oswald, R, Mao, Y. | | Deposit date: | 2022-01-24 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the phosphatidylinositol 3-phosphate binding domain from the Legionella effector SetA
To Be Published
|
|
