6YI3
 
 | |
6YJ0
 
 | |
6YJL
 
 | |
6YP5
 
 | |
6YQ5
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
6YRR
 
 | | Heimdallarchaeota profilin | | Descriptor: | Profilin | | Authors: | Celestine, C. | | Deposit date: | 2020-04-20 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Heimdallarchaea encodes profilin with eukaryotic-like actin regulation and polyproline binding
To Be Published
|
|
6YSE
 
 | |
6YTC
 
 | |
6YTS
 
 | | Solution NMR structure of type-I ribosome-inactivating protein trichobakin (TBK) | | Descriptor: | Trichobakin | | Authors: | Britikov, V.V, Bocharov, E.V, Britikova, E.V, Le, T.B.T, Phan, C.V, Boyko, K.M, Arseniev, A.S, Usanov, S.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of type-I ribosome-inactivating protein trichobakin (TBK)
To Be Published
|
|
6YW8
 
 | |
6YY4
 
 | | Parallel 17-mer DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*AP*AP*GP*GP*GP*TP*GP*GP*GP*A)-3') | | Authors: | Srb, P, Curtis, C, Veverka, V. | | Deposit date: | 2020-05-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Overlapping but distinct: a new model for G-quadruplex biochemical specificity.
Nucleic Acids Res., 49, 2021
|
|
6Z0G
 
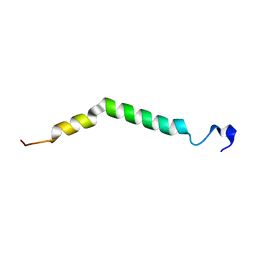 | | Structure of TREM2 transmembrane helix in DPC micelles | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Steiner, A, Schlepkow, K, Brunner, B, Steiner, H, Haass, C, Hagn, F. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | gamma-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics.
Embo J., 39, 2020
|
|
6Z0H
 
 | | Structure of TREM2 transmembrane helix K186A variant in DPC micelles | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Steiner, A, Schlepkow, K, Brunner, B, Steiner, H, Haass, C, Hagn, F. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | gamma-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics.
Embo J., 39, 2020
|
|
6Z0I
 
 | | Structure of the TREM2 transmembrane helix in complex with DAP12 in DPC micelles | | Descriptor: | Triggering receptor expressed on myeloid cells 2 | | Authors: | Steiner, A, Schlepkow, K, Brunner, B, Steiner, H, Haass, C, Hagn, F. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | gamma-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics.
Embo J., 2020
|
|
6Z29
 
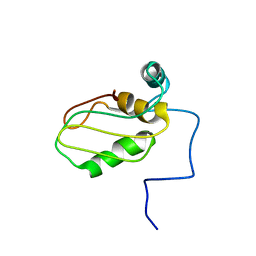 | | Structure of eIF4G1 (37-49) - PUB1 RRM3 chimera in solution | | Descriptor: | Eukaryotic initiation factor 4F subunit p150,Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Chaves-Arquero, B, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | eIF4G1 N-terminal intrinsically disordered domain is a multi-docking station for RNA, Pab1, Pub1, and self-assembly.
Front Mol Biosci, 9, 2022
|
|
6Z35
 
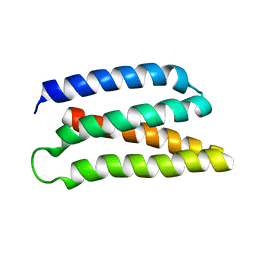 | | De-novo Maquette 2 protein with buried ion-pair | | Descriptor: | Maquette 2-1ip | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M, Asami, S, Mader, S, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Design of buried charged networks in artificial proteins.
Nat Commun, 12, 2021
|
|
6Z40
 
 | |
6Z41
 
 | |
6Z5N
 
 | | DnaJB1 JD-GF | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Avraham-Abayev, M, London, N, Rosenzweig, R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | HSP40 proteins use class-specific regulation to drive HSP70 functional diversity.
Nature, 587, 2020
|
|
6Z98
 
 | | NMR solution structure of the peach allergen Pru p 1.0101 | | Descriptor: | Major allergen Pru p 1 | | Authors: | Eidelpes, R, Fuehrer, S, Hofer, F, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Zeatin Binding of the Peach Allergen Pru p 1 .
J.Agric.Food Chem., 69, 2021
|
|
6ZBI
 
 | | Ternary complex of Calmodulin bound to 2 molecules of NHE1 | | Descriptor: | CALCIUM ION, Calmodulin-1, Sodium/hydrogen exchanger 1 | | Authors: | Prestel, A, Kragelund, B.B, Pedersen, E.S, Pedersen, S.F, Sjoegaard-Frich, L.M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Dynamic Na + /H + exchanger 1 (NHE1) - calmodulin complexes of varying stoichiometry and structure regulate Ca 2+ -dependent NHE1 activation.
Elife, 10, 2021
|
|
6ZDB
 
 | | NMR structural analysis of yeast Cox13 reveals its C-terminus in interaction with ATP | | Descriptor: | Cytochrome c oxidase subunit 13, mitochondrial | | Authors: | Shu, Z, Pontus, P, Peter, B, Lena, M, Pia, A. | | Deposit date: | 2020-06-14 | | Release date: | 2021-05-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural analysis of the yeast cytochrome c oxidase subunit Cox13 and its interaction with ATP.
Bmc Biol., 19, 2021
|
|
6ZFV
 
 | |
6ZL2
 
 | |
6ZL9
 
 | |
