5NIP
 
 | |
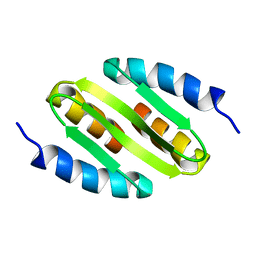
5NIQ
 
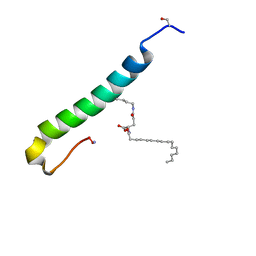 | |
5NKO
 
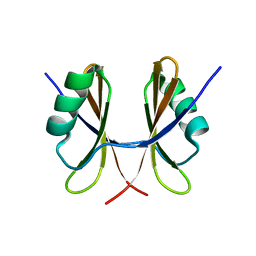 | | Solution structure of the C-terminal domain of S. aureus Hibernating Promoting Factor (CTD-SaHPF) | | Descriptor: | Ribosome hibernation promotion factor | | Authors: | Usachev, K.S, Khusainov, I.S, Ayupov, R.K, Validov, S.Z, Kieffer, B, Yusupov, M.M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structures and dynamics of hibernating ribosomes from Staphylococcus aureus mediated by intermolecular interactions of HPF.
EMBO J., 36, 2017
|
|
5NMY
 
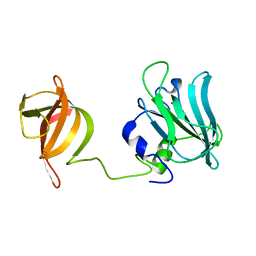 | |
5NOC
 
 | |
5NPA
 
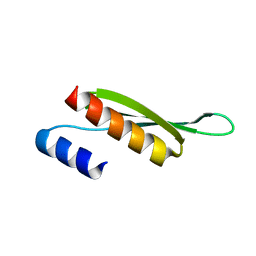 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPG
 
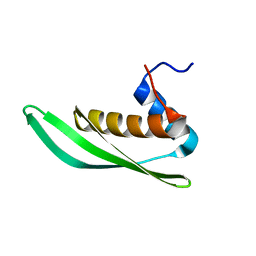 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPN
 
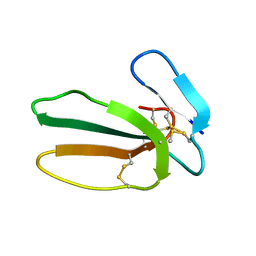 | |
5NQ4
 
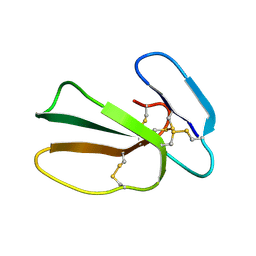 | | Cytotoxin-1 in DPC-micelle | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Volynsky, P.E, Pustovalova, Y.E, Konshina, A.G, Utkin, Y.N, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Impact of membrane partitioning on the spatial structure of an S-type cobra cytotoxin.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5NR5
 
 | |
5NR6
 
 | |
5NVB
 
 | |
5NVP
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | Envelope glycoprotein,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWM
 
 | |
5NWU
 
 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | wtFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWV
 
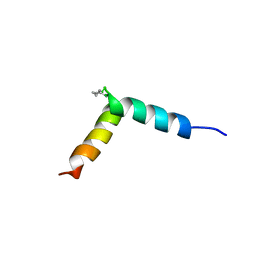 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NWW
 
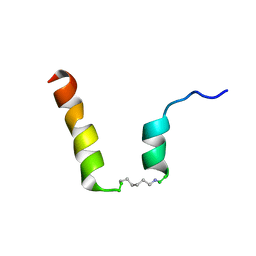 | | NMR assignment and structure of a peptide derived from the fusion peptide of HIV-1 gp41 in the presence of dodecylphosphocholine micelles | | Descriptor: | scrFP-tag,Gp41 | | Authors: | Jimenez, M.A, Serrano, S, Nieva, J.L, Huarte, N. | | Deposit date: | 2017-05-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure-Related Roles for the Conservation of the HIV-1 Fusion Peptide Sequence Revealed by Nuclear Magnetic Resonance.
Biochemistry, 56, 2017
|
|
5NYS
 
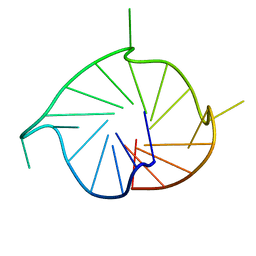 | | M2 G-quadruplex dilute solution | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYT
 
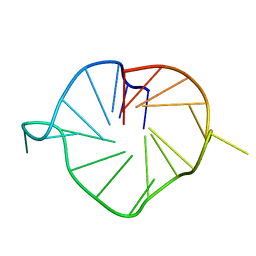 | | M2 G-quadruplex 20 wt% ethylene glycol | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYU
 
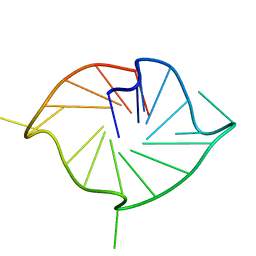 | | M2 G-quadruplex 10 wt% PEG8000 | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NZ9
 
 | | NMR structure of an EphA2-Sam fragment | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The Sam-Sam interaction between Ship2 and the EphA2 receptor: design and analysis of peptide inhibitors.
Sci Rep, 7, 2017
|
|
5O1Q
 
 | | LysF1 sh3b domain structure | | Descriptor: | sh3b domain | | Authors: | Benesik, M, Novacek, J, Janda, L, Dopitova, R, Pernisova, M, Melkova, K, Tisakova, L, Doskar, J, Zidek, L, Hejatko, J, Pantucek, R. | | Deposit date: | 2017-05-19 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity.
Virus Genes, 54, 2018
|
|
5O1T
 
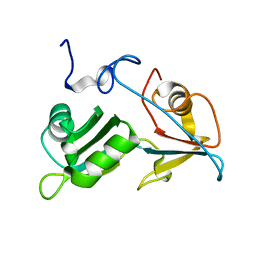 | |
5O2M
 
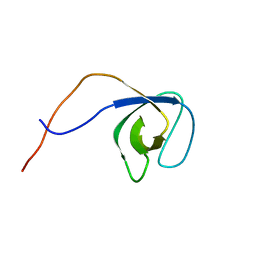 | | p130Cas SH3 domain | | Descriptor: | Breast cancer anti-estrogen resistance 1 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
5O2P
 
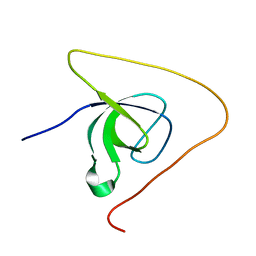 | | p130Cas SH3 domain PTP-PEST peptide chimera | | Descriptor: | Breast cancer anti-estrogen resistance 1,Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Hexnerova, R, Veverka, V. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CAS SH3 domain selectivity and regulation reveals new CAS interaction partners.
Sci Rep, 7, 2017
|
|
