7FBV
 
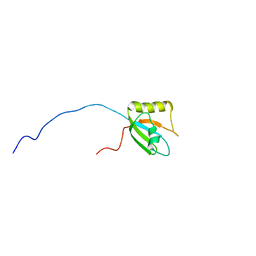 | |
7FGT
 
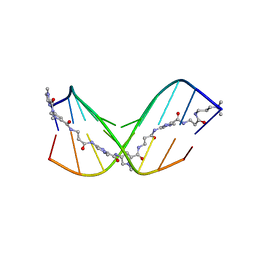 | | Discovery of DS15060524; Gene targeting chimera (GeneTAC) for the treatment of Friedreich's Ataxia (FRDA) | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*AP*GP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*CP*TP*TP*CP*TP*T)-3'), ~{N}-[3-[3-(dimethylamino)propylamino]-3-oxidanylidene-propyl]-1-methyl-4-[3-[[1-methyl-4-[[1-methyl-4-[3-[[1-methyl-4-[(1-methylimidazol-2-yl)carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]imidazol-2-yl]carbonylamino]pyrrol-2-yl]carbonylamino]propanoylamino]imidazole-2-carboxamide | | Authors: | Takase, N, Kawai, G, Igarashi, W, Katagiri, T. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery of DS15060524; Gene targeting chimera (GeneTAC) for the treatment of Friedreich's Ataxia (FRDA)
To be published
|
|
7FHI
 
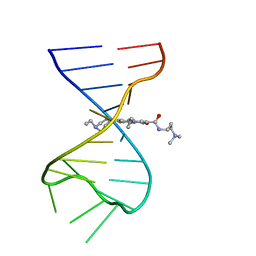 | |
7FHQ
 
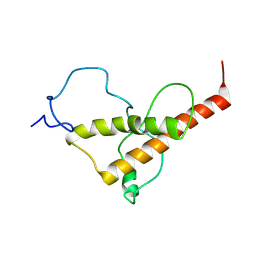 | |
7FJ0
 
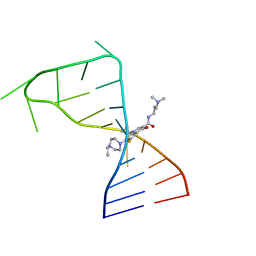 | |
7JGI
 
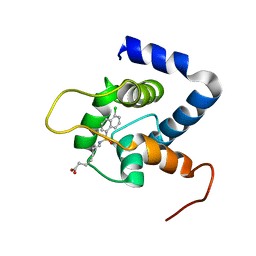 | | NMR structure of the cNTnC-cTnI chimera bound to A7 | | Descriptor: | 7-{[(5-chloronaphthalen-1-yl)sulfonyl]amino}heptanoic acid, CALCIUM ION, Troponin C, ... | | Authors: | Cai, F, Robertson, I.M, Kampourakis, T, Klein, B.A, Sykes, B.D. | | Deposit date: | 2020-07-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Role of Electrostatics in the Mechanism of Cardiac Thin Filament Based Sensitizers.
Acs Chem.Biol., 15, 2020
|
|
7JGX
 
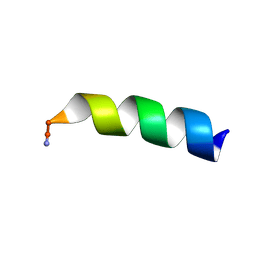 | |
7JGY
 
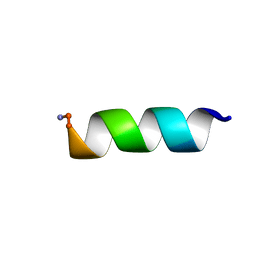 | |
7JH1
 
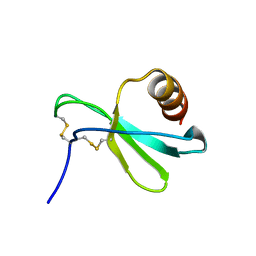 | |
7JHF
 
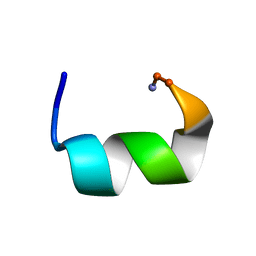 | |
7JI1
 
 | |
7JK8
 
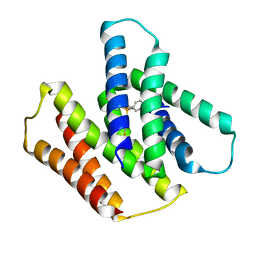 | | EmrE S64V mutant bound to tetra(4-fluorophenyl)phosphonium at pH 5.8 | | Descriptor: | Multidrug SMR transporter, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Hisao, G, Mandala, V.S, Thomas, N.E, Soltani, M, Salter, E.A, Davis Jr, J.H, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers.
Nat Commun, 12, 2021
|
|
7JMY
 
 | |
7JN6
 
 | |
7JNN
 
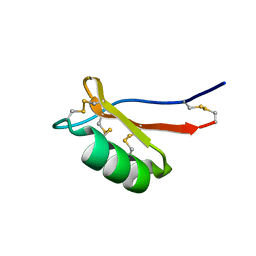 | |
7JPM
 
 | |
7JQ8
 
 | | Solution NMR structure of human Brd3 ET domain | | Descriptor: | Bromodomain-containing protein 3, Integrase peptide | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, J.M, Montelione, G.T. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JS6
 
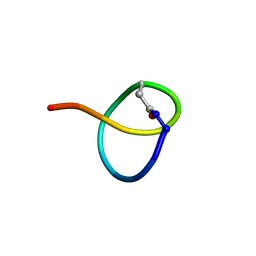 | | Solution NMR structure of des-citrulassin F | | Descriptor: | des-citrulassin F | | Authors: | Harris, L.A, Mitchell, D.A. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Reactivity-Based Screening for Citrulline-Containing Natural Products Reveals a Family of Bacterial Peptidyl Arginine Deiminases.
Acs Chem.Biol., 15, 2020
|
|
7JSQ
 
 | | Refined structure of the C-terminal domain of DNAJB6b | | Descriptor: | DnaJ homolog subfamily B member 6 | | Authors: | Karamanos, T.K, Clore, G.M. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An S/T motif controls reversible oligomerization of the Hsp40 chaperone DNAJB6b through subtle reorganization of a beta sheet backbone.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JU1
 
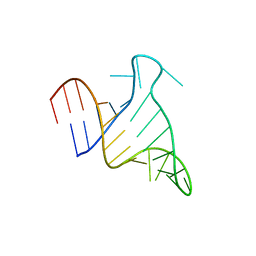 | | The FARFAR-NMR Ensemble of 29-mer HIV-1 Trans-activation Response Element RNA (N=20) | | Descriptor: | RNA (29-MER) | | Authors: | Shi, H, Rangadurai, A, Roy, R, Yesselman, J.D, Al-Hashimi, H.M. | | Deposit date: | 2020-08-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rapid and accurate determination of atomistic RNA dynamic ensemble models using NMR and structure prediction
Nat Commun, 11, 2020
|
|
7JU9
 
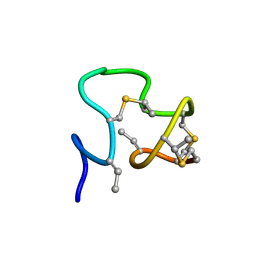 | |
7JVF
 
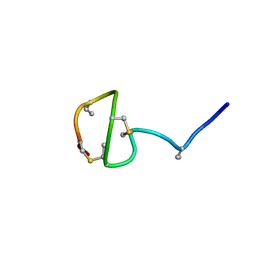 | |
7JXG
 
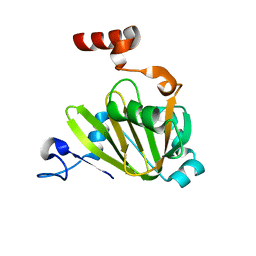 | | Structural model for Fe-containing human acireductone dioxygenase | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, FE (II) ION | | Authors: | Pochapsky, T.C, Liu, X, Deshpande, A, Ringe, D, Garber, A, Ryan, J. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Model for the Solution Structure of Human Fe(II)-Bound Acireductone Dioxygenase and Interactions with the Regulatory Domain of Matrix Metalloproteinase I (MMP-I).
Biochemistry, 59, 2020
|
|
7JYN
 
 | | Solution NMR structure of human Brd3 ET complexed with NSD3(148-184) peptide | | Descriptor: | Bromodomain-containing protein 3, Histone-lysine N-methyltransferase NSD3 | | Authors: | Aiyer, S, Swapna, G.V.T, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
7JYZ
 
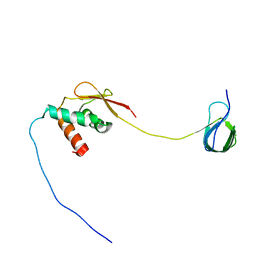 | | Solution NMR structure and dynamics of human Brd3 ET in complex with MLV IN CTD | | Descriptor: | Bromodomain-containing protein 3, Integrase | | Authors: | Aiyer, S, Liu, G, Swapna, G.V.T, Hao, J, Ma, L.C, Roth, M.J, Montelione, G.T. | | Deposit date: | 2020-09-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A common binding motif in the ET domain of BRD3 forms polymorphic structural interfaces with host and viral proteins.
Structure, 29, 2021
|
|
