7B3K
 
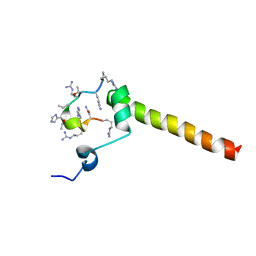 | | Dynamic complex between all-D-enantiomeric peptide D3 with L723P mutant of amyloid precursor protein (APP) 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
7B4Z
 
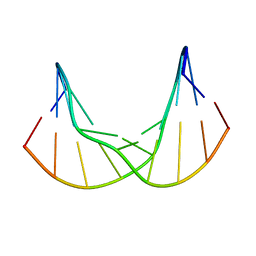 | | Synthetic DNA duplex dodecamer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*TP*G)-3') | | Authors: | Lomzov, A.A, Shernuykov, A.V, Sviridov, E.A, Shevelev, G.Y, Bagryanskaya, E.G, Pyshnyi, D.V. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Study of a DNA Duplex by Nuclear Magnetic Resonance and Molecular Dynamics Simulations. Validation of Pulsed Dipolar Electron Paramagnetic Resonance Distance Measurements Using Triarylmethyl-Based Spin Labels.
J Phys Chem B, 120, 2016
|
|
7B71
 
 | | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(SGT)P*G)-3') | | Authors: | Lomzov, A.A, Shernuykov, A.V, Apukhtina, V.S, Pyshnyi, D.V. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Single modified phosphoryl guanidine DNA duplex, Sp diastereomer
To Be Published
|
|
7B72
 
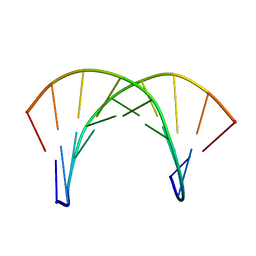 | | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*CP*CP*GP*CP*TP*G)-3'), DNA (5'-D(*CP*AP*GP*CP*GP*GP*CP*GP*(RGT)P*G)-3') | | Authors: | Lomzov, A.A, Pyshnyi, D.V, Shernuykov, A.V, Apukhtina, V.S. | | Deposit date: | 2020-12-09 | | Release date: | 2021-01-13 | | Last modified: | 2023-08-23 | | Method: | SOLUTION NMR | | Cite: | DNA duplex with phosphoryl guanidine moiety, Rp-diastereomer
To Be Published
|
|
7B7O
 
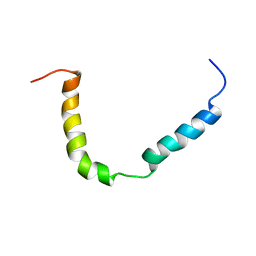 | | Solution structure of A. thaliana core TatA in DHPC micelles | | Descriptor: | Sec-independent protein translocase protein TATA, chloroplastic | | Authors: | Pettersson, P, Ye, W, Jakob, M, Tannert, F, Klosgen, R.B, Maler, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-01-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of plant TatA in micelles and lipid bilayers studied by solution NMR.
FEBS J, 285, 2018
|
|
7B9X
 
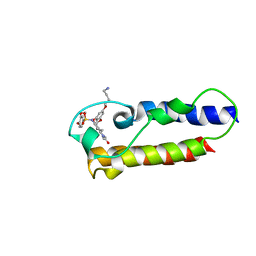 | | NMR2 structure of TRIM24-BD in complex with a precursor of IACS-9571 | | Descriptor: | N-{6-[3-(4-Aminobutoxy)-5-propoxyphenoxy]-1,3-dimethyl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl}-3,4-dimethoxybenzene-1-sulfonamide, Transcription intermediary factor 1-alpha | | Authors: | Orts, J, Torres, F, Milbradt, A.G, Walser, R. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Molecular Replacement Provides New Insights into Binding Modes to Bromodomains of BRD4 and TRIM24.
J.Med.Chem., 65, 2022
|
|
7BBB
 
 | |
7BCJ
 
 | |
7BEV
 
 | |
7BFS
 
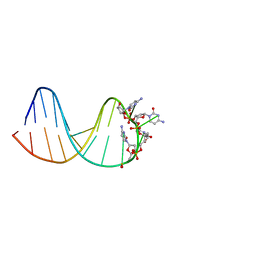 | | deoxyxylose nucleic acid hairpin | | Descriptor: | DNA (5'-D(*AP*GP*CP*AP*AP*TP*CP*CP*(XC)P*(XC)P*(XC)P*(XC)P*GP*GP*AP*TP*TP*GP*CP*T)-3') | | Authors: | Mattelaer, C.-A, Mohitosh, M, Smets, L, Maiti, M, Schepers, G, Mattelaer, H.-P, Rosemeyer, H, Herdewijn, P, Lescrinier, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stable Hairpin Structures Formed by Xylose-Based Nucleic Acids.
Chembiochem, 22, 2021
|
|
7BFX
 
 | | deoxyxylose nucleic acid hairpin | | Descriptor: | dXyNA (5'-D(*(XA)P*(XG)P*(XC)P*(XA)P*(XA)P*(XT)P*(XC)P*(XC)P*(XC)P*(XC)P*(XC)P*(XC)P*(XG)P*(XG)P*(XA)P*(XT)P*(XT)P*(XG)P*(XC)P*T)-3') | | Authors: | Mattelaer, C.-A, Mohitosh, M, Smets, L, Maiti, M, Schepers, G, Mattelaer, H.-P, Rosemeyer, H, Herdewijn, P, Lescrinier, E. | | Deposit date: | 2021-01-05 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-31 | | Method: | SOLUTION NMR | | Cite: | Stable Hairpin Structures Formed by Xylose-Based Nucleic Acids.
Chembiochem, 22, 2021
|
|
7BGH
 
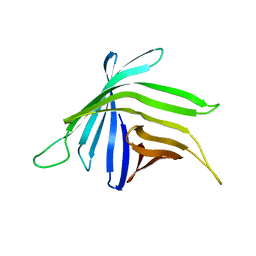 | |
7BI0
 
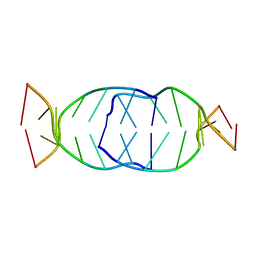 | | GA repetition with i-motif clip at 5'-end | | Descriptor: | (CH+)C(CH+)GAGA, C(CH+)CGAGA | | Authors: | Novotny, A, Novotny, J. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing structural peculiarities of homopurine GA repetition stuck by i-motif clip.
Nucleic Acids Res., 49, 2021
|
|
7BL0
 
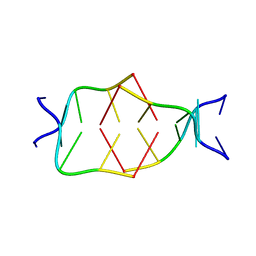 | | GA attached to an i-motif clip at 3'-end | | Descriptor: | GA(CH+)C(5MeCH+), GAC(HCY)(5MeC) | | Authors: | Novotny, A, Novotny, J. | | Deposit date: | 2021-01-17 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Revealing structural peculiarities of homopurine GA repetition stuck by i-motif clip.
Nucleic Acids Res., 49, 2021
|
|
7BLM
 
 | |
7BMA
 
 | |
7BPL
 
 | | Solution NMR structure of NF1; de novo designed protein with a novel fold | | Descriptor: | NF1 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | Descriptor: | NF2 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPN
 
 | | Solution NMR structure of NF7; de novo designed protein with a novel fold | | Descriptor: | NF7 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BPP
 
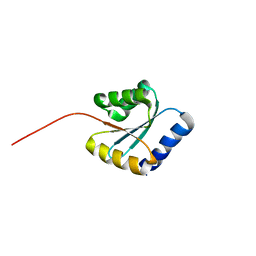 | | Solution NMR structure of NF5; de novo designed protein with a novel fold | | Descriptor: | NF5 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQB
 
 | | Solution NMR structure of NF6; de novo designed protein with a novel fold | | Descriptor: | NF6 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQC
 
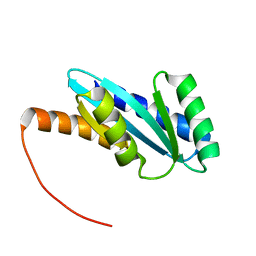 | | Solution NMR structure of NF4; de novo designed protein with a novel fold | | Descriptor: | NF4 | | Authors: | Kobayashi, N, Nagashima, T, Minami, S, Koga, R, Chikenji, T, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
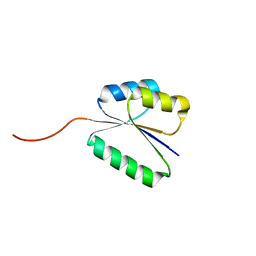 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | Descriptor: | NF8 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQE
 
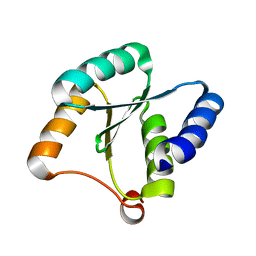 | | Solution NMR structure of NF3; de novo designed protein with a novel fold | | Descriptor: | NF3 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQM
 
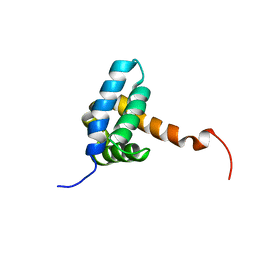 | | Solution NMR structure of fold-0 Chantal; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Chantal | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
