4ASW
 
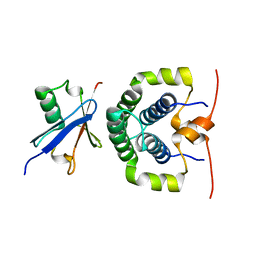 | | Structure of the complex between the N-terminal dimerisation domain of Sgt2 and the UBL domain of Get5 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2, UBIQUITIN-LIKE PROTEIN MDY2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AXP
 
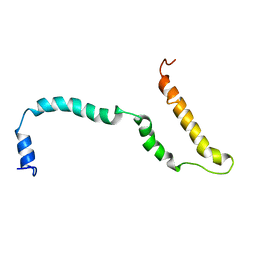 | | NMR structure of Hsp12, a protein induced by and required for dietary restriction-induced lifespan extension in yeast. | | Descriptor: | 12 KDA HEAT SHOCK PROTEIN | | Authors: | Herbert, A.P, Riesen, M, Bloxam, L, Kosmidou, E, Wareing, B.M, Johnson, J.R, Phelan, M.M, Pennington, S.R, Lian, L.Y, Morgan, A. | | Deposit date: | 2012-06-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Hsp12, a Protein Induced by and Required for Dietary Restriction-Induced Lifespan Extension in Yeast.
Plos One, 7, 2012
|
|
4B19
 
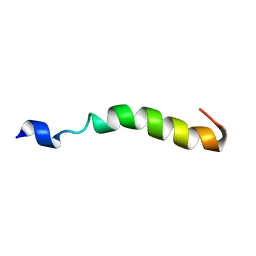 | |
4B1Q
 
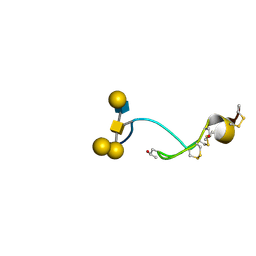 | | NMR structure of the glycosylated conotoxin CcTx from Conus consors | | Descriptor: | CONOTOXIN CCTX, alpha-L-galactopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-6)]2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Hocking, H.G, Gerwig, G.J, Favreau, P, Stocklin, R, Kamerling, J.P, Boelens, R. | | Deposit date: | 2012-07-12 | | Release date: | 2013-02-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the O-Glycosylated Conopeptide Cctx from Conus Consors Venom.
Chemistry, 19, 2013
|
|
4B2R
 
 | | Solution structure of CCP modules 10-11 of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Makou, E, Mertens, H.D.T, Maciejewski, M, Soares, D.C, Matis, I, Schmidt, C.Q, Herbert, A.P, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-10 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ccp Modules 10-12 Illuminates Functional Architecture of the Complement Regulator, Factor H.
J.Mol.Biol., 424, 2012
|
|
4B2S
 
 | | Solution structure of CCP modules 11-12 of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Makou, E, Mertens, H.D, Maciejewski, M, Soares, D.C, Matis, I, Schmidt, C.Q, Herbert, A.P, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ccp Modules 10-12 Illuminates Functional Architecture of the Complement Regulator, Factor H.
J.Mol.Biol., 424, 2012
|
|
4B2U
 
 | | S67, A spider venom toxin peptide from Sicarius dolichocephalus | | Descriptor: | S67 | | Authors: | Loening, N.M, Wilson, Z.N, Zobel-Thropp, P.A, Binford, G.J. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2013-02-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Two Homologous Venom Peptides from Sicarius Dolichocephalus.
Plos One, 8, 2013
|
|
4B2V
 
 | | S64, a spider venom toxin peptide from Sicarius dolichocephalus | | Descriptor: | S64 | | Authors: | Loening, N.M, Wilson, Z.N, Zobel-Thropp, P.A, Binford, G.J. | | Deposit date: | 2012-07-18 | | Release date: | 2013-01-16 | | Last modified: | 2013-02-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Two Homologous Venom Peptides from Sicarius Dolichocephalus
Plos One, 8, 2013
|
|
4B6U
 
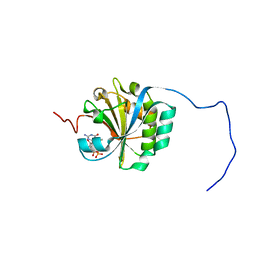 | | Solution structure of eIF4E3 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 3 | | Authors: | Osborne, M.J, Volpon, L, Kornblatt, J.A, Culkjovic-Kraljcic, B, Baguet, A, Borden, K.L.B. | | Deposit date: | 2012-08-15 | | Release date: | 2013-03-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Eif4E3 Acts as a Tumor Suppressor by Utilizing an Atypical Mode of Methyl-7-Guanosine CAP Recognition
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4B6V
 
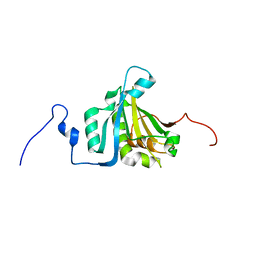 | | The third member of the eIF4E family represses gene expression via a novel mode of recognition of the methyl-7 guanosine cap moiety | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TYPE 3 | | Authors: | Osborne, M.J, Volpon, L, Kornblatt, J.A, Culjkovic-Kraljcic, B, Baguet, A, Borden, K.L.B. | | Deposit date: | 2012-08-15 | | Release date: | 2013-02-27 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Eif4E3 Acts as a Tumor Suppressor by Utilizing an Atypical Mode of Methyl-7-Guanosine CAP Recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4B8T
 
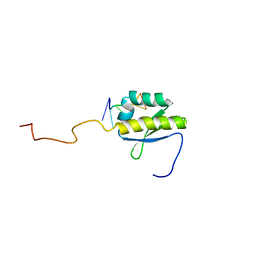 | | RNA BINDING PROTEIN Solution structure of the third KH domain of KSRP in complex with the G-rich target sequence. | | Descriptor: | 5'-R(*AP*GP*GP*GP*UP)-3', KH-TYPE SPLICING REGULATORY PROTEIN | | Authors: | Nicastro, G, Garcia-Mayoral, M.F, Hollingworth, D, Kelly, G, Martin, S.R, Briata, P, Gherzi, R, Ramos, A. | | Deposit date: | 2012-08-30 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Noncanonical G Recognition Mediates Ksrp Regulation of Let-7 Biogenesis
Nat.Struct.Mol.Biol., 19, 2012
|
|
4BA8
 
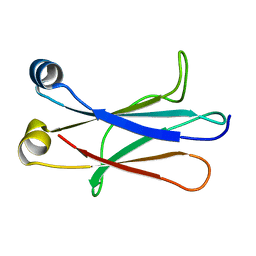 | | High resolution NMR structure of the C mu3 domain from IgM | | Descriptor: | IG MU CHAIN C REGION SECRETED FORM | | Authors: | Mueller, R, Kern, T, Graewert, M.A, Madl, T, Peschek, J, Groll, M, Sattler, M, Buchner, J. | | Deposit date: | 2012-09-12 | | Release date: | 2013-06-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High Resolution Structures of the Igm Fc Domains Reveal Principles of its Hexamer Formation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BD3
 
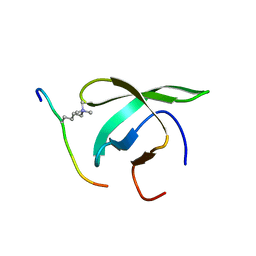 | |
4BEH
 
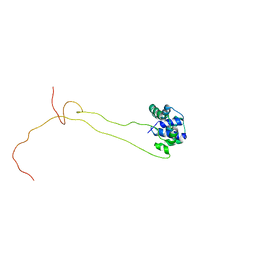 | | Solution structure of human ribosomal protein P1.P2 heterodimer | | Descriptor: | 60S ACIDIC RIBOSOMAL PROTEIN P1, 60S ACIDIC RIBOSOMAL PROTEIN P2 | | Authors: | Lee, K.M, Yusa, K, Chu, L.O, Wing-Heng Yu, C, Shaw, P.C, Oono, M, Miyoshi, T, Ito, K, Wong, K.B, Uchiumi, T. | | Deposit date: | 2013-03-10 | | Release date: | 2013-08-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human P1P2 Heterodimer Provides Insights Into the Role of Eukaryotic Stalk in Recruiting the Ribosome-Inactivating Protein Trichosanthin to the Ribosome.
Nucleic Acids Res., 41, 2013
|
|
4BF8
 
 | | Fpr4 PPI domain | | Descriptor: | FPR4 | | Authors: | Monneau, Y, Mackereth, C. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and Activity of the Peptidyl-Prolyl Isomerase Domain from the Histone Chaperone Fpr4 Towards Histone H3 Proline Isomerization
J.Biol.Chem., 288, 2013
|
|
4BH9
 
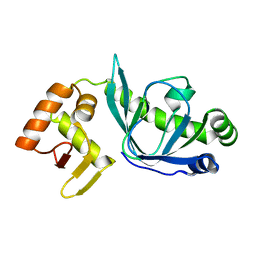 | |
4BHP
 
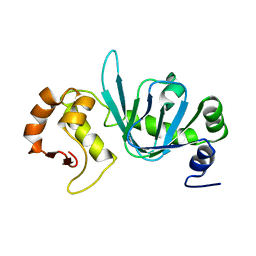 | |
4BIT
 
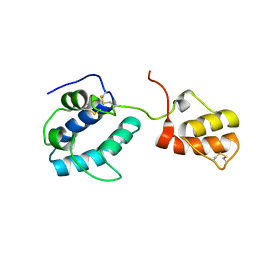 | | solution structure of cerebral dopamine neurotrophic factor (CDNF) | | Descriptor: | CEREBRAL DOPAMINE NEUROTROPHIC FACTOR | | Authors: | Latge, C, Cabral, K.M.S, Raymundo, D.P, Foguel, D, Herrmann, T, Almeida, M.S. | | Deposit date: | 2013-04-13 | | Release date: | 2014-04-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and Dynamics of Full-Length Human Cerebral Dopamine Neurotrophic Factor and its Neuroprotective Role Against Alpha-Synuclein Oligomers.
J.Biol.Chem., 290, 2015
|
|
4BMF
 
 | |
4BS2
 
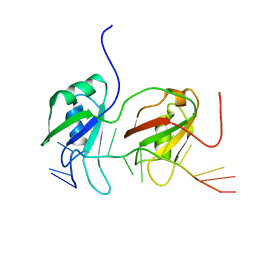 | | NMR structure of human TDP-43 tandem RRMs in complex with UG-rich RNA | | Descriptor: | 5'-R(*GP*UP*GP*UP*GP*AP*AP*UP*GP*AP*AP*UP)-3', TAR DNA-BINDING PROTEIN 43 | | Authors: | Lukavsky, P.J, Daujotyte, D, Tollervey, J.R, Ule, J, Stuani, C, Buratti, E, Baralle, F.E, Damberger, F.F, Allain, F.H.T. | | Deposit date: | 2013-06-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of Ug-Rich RNA Recognition by the Human Splicing Factor Tdp-43
Nat.Struct.Mol.Biol., 20, 2013
|
|
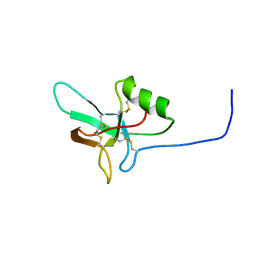
4BWH
 
 | |
4BXL
 
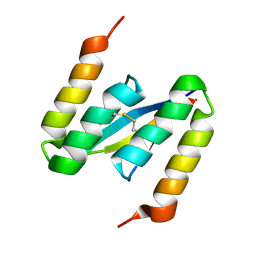 | | Structure of alpha-synuclein in complex with an engineered binding protein | | Descriptor: | ALPHA SYNUCLEIN, AS69 | | Authors: | Mirecka, E.A, Shaykhalishahi, H, Lecher, J, Stoldt, M, Hoyer, W. | | Deposit date: | 2013-07-12 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Sequestration of a Beta-Hairpin for Control of Alpha-Synuclein Aggregation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4BXU
 
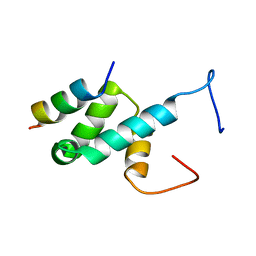 | | Structure of Pex14 in complex with Pex5 LVxEF motif | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Kooshapur, H, Meyer, H.N, Madl, T, Sattler, M. | | Deposit date: | 2013-07-15 | | Release date: | 2013-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Novel Pex14 Interacting Site of Human Pex5 is Critical for Matrix Protein Import Into Peroxisomes.
J.Biol.Chem., 289, 2014
|
|
4BY9
 
 | | The structure of the Box CD enzyme reveals regulation of rRNA methylation | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*CP*AP*UP*CP*AP*CP)-3', 50S RIBOSOMAL PROTEIN L7AE, FIBRILLARIN-LIKE RRNA/TRNA 2'-O-METHYLTRANSFERASE, ... | | Authors: | Lapinaite, A, Simon, B, Skjaerven, L, Rakwalska-Bange, M, Gabel, F, Carlomagno, T. | | Deposit date: | 2013-07-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Box C/D Enzyme Reveals Regulation of RNA Methylation.
Nature, 502, 2013
|
|
4BYA
 
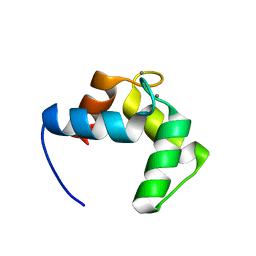 | | Calmodulin, C-terminal domain, M144H mutant | | Descriptor: | CALCIUM ION, CALMODULIN, C-TERMINAL DOMAIN, ... | | Authors: | Moroz, Y.S, Wu, Y, van Nuland, N.A.J, Korendovych, I.V. | | Deposit date: | 2013-07-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | New Tricks for Old Proteins: Single Mutations in a Nonenzymatic Protein Give Rise to Various Enzymatic Activities.
J.Am.Chem.Soc., 137, 2015
|
|
