2N63
 
 | | Structure of C4VG16KRKP | | Descriptor: | PENTANOIC ACID, antimicrobial peptide C4VG16KRKP | | Authors: | Bhunia, A, Datta, A. | | Deposit date: | 2015-08-11 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Designing potent antimicrobial peptides by disulphide linked dimerization and N-terminal lipidation to increase antimicrobial activity and membrane perturbation: Structural insights into lipopolysaccharide binding.
J Colloid Interface Sci, 461, 2016
|
|
2N64
 
 | |
2N65
 
 | | Disulphide linked homodimer of designed antimicrobial peptide VG16KRKP | | Descriptor: | antimicrobial peptide VG16KRKP | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2015-08-12 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Designing potent antimicrobial peptides by disulphide linked dimerization and N-terminal lipidation to increase antimicrobial activity and membrane perturbation: Structural insights into lipopolysaccharide binding.
J Colloid Interface Sci, 461, 2016
|
|
2N66
 
 | |
2N67
 
 | |
2N68
 
 | |
2N69
 
 | |
2N6A
 
 | |
2N6B
 
 | |
2N6C
 
 | |
2N6D
 
 | |
2N6E
 
 | |
2N6F
 
 | | Structure of Pleiotrophin | | Descriptor: | Pleiotrophin | | Authors: | Ryan, E.O, Shen, D, Wang, X. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural studies reveal an important role for the pleiotrophin C-terminus in mediating interactions with chondroitin sulfate.
Febs J., 283, 2016
|
|
2N6G
 
 | |
2N6H
 
 | |
2N6I
 
 | |
2N6J
 
 | | Solution structure of Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile | | Descriptor: | ZINC ION, Zinc metalloprotease Zmp1 | | Authors: | Banci, L, Cantini, F, Scarselli, M, Rubino, J.T, Martinelli, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of zinc-bound Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile.
J.Biol.Inorg.Chem., 21, 2016
|
|
2N6L
 
 | | Solution NMR structure of Outer Membrane Protein G from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprG | | Authors: | Kucharska, I, Seelheim, P, Edrington, T.C, Liang, B, Tamm, L.K. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | OprG Harnesses the Dynamics of its Extracellular Loops to Transport Small Amino Acids across the Outer Membrane of Pseudomonas aeruginosa.
Structure, 23, 2015
|
|
2N6M
 
 | |
2N6N
 
 | | Structure Determination for spider toxin, U4-agatoxin-Ao1a | | Descriptor: | U4-agatoxin-Ao1a | | Authors: | Pineda, S.S, Chin, Y.K.-Y, Mobli, M.S, King, G.F. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural venomics reveals evolution of a complex venom by duplication and diversification of an ancient peptide-encoding gene.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2N6O
 
 | |
2N6P
 
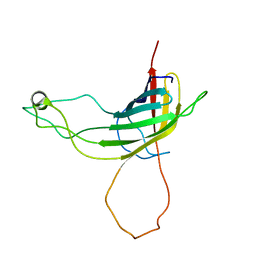 | | Solution NMR structure of Outer Membrane Protein G P92A mutant from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprG | | Authors: | Kucharska, I, Seelheim, P, Edrington, T.C, Liang, B, Tamm, L.K. | | Deposit date: | 2015-08-27 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | OprG Harnesses the Dynamics of its Extracellular Loops to Transport Small Amino Acids across the Outer Membrane of Pseudomonas aeruginosa.
Structure, 23, 2015
|
|
2N6R
 
 | |
2N6S
 
 | |
2N6T
 
 | |
