2MZY
 
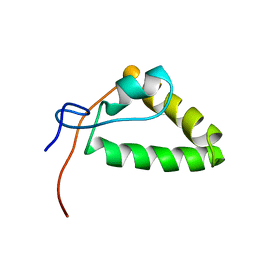 | | 1H, 13C, and 15N Chemical Shift Assignments and structure of Probable Fe(2+)-trafficking protein from Burkholderia pseudomallei 1710b. | | Descriptor: | Probable Fe(2+)-trafficking protein | | Authors: | Tang, C, Yang, F, Barnwal, R, Varani, G, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Probable Fe(2+)-trafficking protein
To be Published
|
|
2MZZ
 
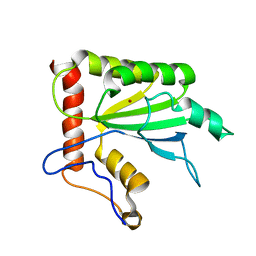 | | NMR structure of APOBEC3G NTD variant, sNTD | | Descriptor: | Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G variant, ZINC ION | | Authors: | Kouno, T, Luengas, E.M, Shigematu, M, Shandilya, S.M.D, Zhang, J, Chen, L, Hara, M, Schiffer, C.A, Harris, R.S, Matsuo, H. | | Deposit date: | 2015-02-28 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2N00
 
 | | NMR Solution structure of AIM2 PYD from Mus musculus | | Descriptor: | Interferon-inducible protein AIM2 | | Authors: | Hou, X, Niu, X. | | Deposit date: | 2015-03-01 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of AIM2 PYD domain from Mus musculus reveals a distinct alpha 2-alpha 3 helix conformation from its human homologues
Biochem.Biophys.Res.Commun., 461, 2015
|
|
2N01
 
 | | NMR structure of VirB9 C-terminal domain in complex with VirB7 N-terminal domain from Xanthomonas citri's T4SS | | Descriptor: | VirB7 protein, VirB9 protein | | Authors: | Oliveira, L.C, Souza, D.P, Salinas, R.K, Wienk, H, Boelens, R, Farah, S.C. | | Deposit date: | 2015-03-03 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | VirB7 and VirB9 Interactions Are Required for the Assembly and Antibacterial Activity of a Type IV Secretion System.
Structure, 24, 2016
|
|
2N02
 
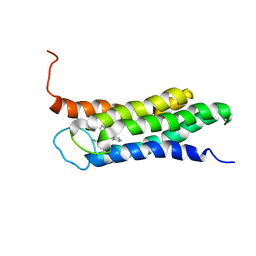 | | Solution structure of the A147T variant of the mitochondrial translocator protein (tspo) in complex with pk11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Integrity of the A147T Polymorph of Mammalian TSPO.
Chembiochem, 16, 2015
|
|
2N03
 
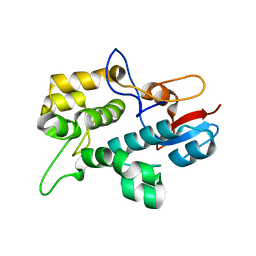 | | Solution NMR Structure plectin repeat domain 6 (4403-4606) of Plectin from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR6354E | | Descriptor: | Plectin | | Authors: | Pulavarti, S, Eletsky, A, Huang, Y, Janjua, H, Xiao, R, Acton, T, Everett, J, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-03-04 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure plectin repeat domain 6 (4403-4606) of Plectin from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR6354E
To be Published
|
|
2N04
 
 | |
2N05
 
 | |
2N06
 
 | | Mdmx-298 | | Descriptor: | 4-[[(4S,5R)-5-(4-chlorophenyl)-4-(3-methoxyphenyl)-2-(4-methoxy-2-propan-2-yloxy-phenyl)-4,5-dihydroimidazol-1-yl]carbonyl]piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2N07
 
 | | Design of a Highly Stable Disulfide-Deleted Mutant of Analgesic Cyclic alpha-Conotoxin Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Yu, R, Seymour, V, Berecki, G, Jia, X, Akcan, M, Adams, D, Kaas, Q, Craik, D. | | Deposit date: | 2015-03-04 | | Release date: | 2016-04-13 | | Method: | SOLUTION NMR | | Cite: | Design of a Highly Stable Disulfide-Deleted Mutant of Analgesic Cyclic alpha-Conotoxin Vc1.1.
To be Published
|
|
2N08
 
 | | NMR structure of a short hydrophobic 11mer peptide in 25 mM SDS solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N09
 
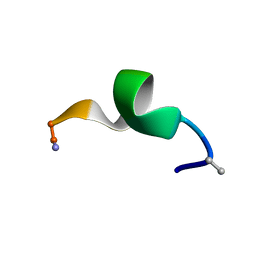 | | NMR structure of a short hydrophobic 11mer peptide in DMSO-d6/H2O (1:3) solution | | Descriptor: | Short hydrophobic peptide with cyclic constraints | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-04 | | Release date: | 2015-04-15 | | Last modified: | 2015-05-27 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N0A
 
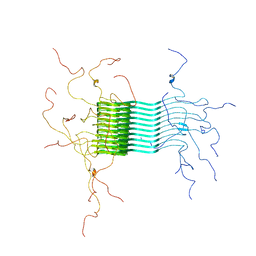 | | Atomic-resolution structure of alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Tuttle, M.D, Comellas, G, Nieuwkoop, A.J, Covell, D.J, Berthold, D.A, Kloepper, K.D, Courtney, J.M, Kim, J.K, Schwieters, C.D, Lee, V.M, George, J.M, Rienstra, C.M. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2N0B
 
 | | NMR structure of Neuromedin C in aqueous solution | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0C
 
 | | NMR structure of Neuromedin C in 10% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0D
 
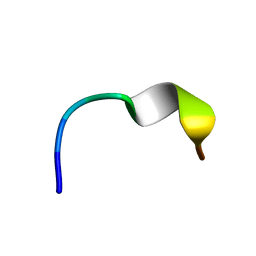 | | NMR structure of Neuromedin C in 25% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0E
 
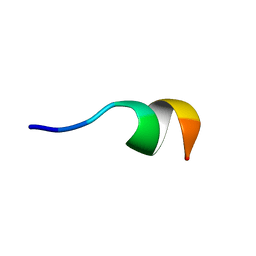 | | NMR structure of Neuromedin C in 40% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0F
 
 | | NMR structure of Neuromedin C in 60% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0G
 
 | | NMR structure of Neuromedin C in 90% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0H
 
 | | NMR structure of Neuromedin C in presence of SDS micelles | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
2N0I
 
 | | NMR solution structure for di-sulfide 11mer peptide | | Descriptor: | di-sulfide 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
2N0J
 
 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostamycin complex | | Descriptor: | RIBOSTAMYCIN, RNA_(27-MER) | | Authors: | Duchardt-Ferner, E, Gottstein-Schmidtke, S.R, Weigand, J.E, Ohlenschlaeger, O.E, Wurm, J, Hammann, C, Suess, B, Woehnert, J. | | Deposit date: | 2015-03-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2N0K
 
 | | Chemical shift assignments and structure of the alpha-crystallin domain from human, HSPB5 | | Descriptor: | Alpha-crystallin B chain | | Authors: | Rajagopal, P, Klevit, R.E, Shi, L, Baker, D. | | Deposit date: | 2015-03-09 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis.
Elife, 4, 2015
|
|
2N0M
 
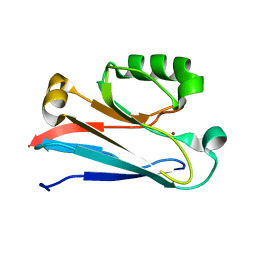 | | The solution structure of the soluble form of the Lipid-modified Azurin from Neisseria gonorrhoeae | | Descriptor: | COPPER (I) ION, Lipid modified azurin protein | | Authors: | Pauleta, S.R, Matzapetakis, M.F, Nobrega, C.F, Carreira, C, Saraiva, I.H. | | Deposit date: | 2015-03-10 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the soluble form of the lipid-modified azurin from Neisseria gonorrhoeae, the electron donor of cytochrome c peroxidase.
Biochim.Biophys.Acta, 1857, 2016
|
|
2N0N
 
 | | NMR solution structure for lactam (5,9) 11mer | | Descriptor: | lactam (5,9) 11mer peptide | | Authors: | Hoang, H.N, Song, K, Hill, T.A, Derksen, D.R, Edmonds, D.J, Kok, W.M, Limberakis, C, Liras, S, Loria, P.M, Mascitti, V, Mathiowetz, A.M, Mitchell, J.M, Piotrowski, D.W, Price, D.A, Stanton, R.V, Suen, J.Y, Withka, J.M, Griffith, D.A, Fairlie, D.P. | | Deposit date: | 2015-03-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Short Hydrophobic Peptides with Cyclic Constraints Are Potent Glucagon-like Peptide-1 Receptor (GLP-1R) Agonists.
J.Med.Chem., 58, 2015
|
|
