1ZPX
 
 | |
1ZQ3
 
 | | NMR Solution Structure of the Bicoid Homeodomain Bound to the Consensus DNA Binding Site TAATCC | | Descriptor: | 5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3', Homeotic bicoid protein | | Authors: | Baird-Titus, J.M, Rance, M, Clark-Baldwin, K, Ma, J, Vrushank, D. | | Deposit date: | 2005-05-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the native K50 Bicoid homeodomain bound to the consensus TAATCC DNA-binding site.
J.Mol.Biol., 356, 2006
|
|
1ZR7
 
 | |
1ZR9
 
 | |
1ZRI
 
 | |
1ZRJ
 
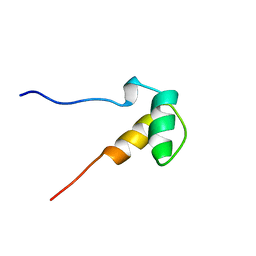 | | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c | | Descriptor: | E1B-55kDa-associated protein 5 isoform c | | Authors: | Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAP domain of human E1B-55kDa-associated protein 5 isoform c
To be Published
|
|
1ZRR
 
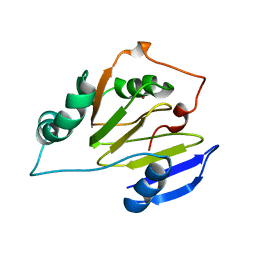 | | Residual Dipolar Coupling Refinement of Acireductone Dioxygenase from Klebsiella | | Descriptor: | E-2/E-2' protein, NICKEL (II) ION | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Ju, T, Hoefler, C, Liang, J. | | Deposit date: | 2005-05-19 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A refined model for the structure of acireductone dioxygenase from Klebsiella ATCC 8724 incorporating residual dipolar couplings
J.Biomol.NMR, 34, 2006
|
|
1ZRY
 
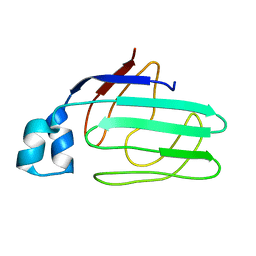 | | NMR structural analysis of apo chicken liver bile acid binding protein | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Ragona, L, Catalano, M, Luppi, M, Cicero, D, Eliseo, T, Foote, J, Fogolari, F, Zetta, L, Molinari, H. | | Deposit date: | 2005-05-23 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Dynamic Studies Suggest that Allosteric Activation Regulates Ligand Binding in Chicken Liver Bile Acid-binding Protein
J.Biol.Chem., 281, 2006
|
|
1ZS5
 
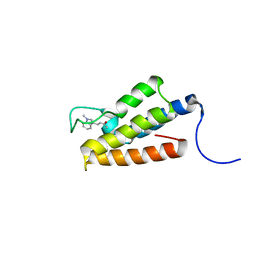 | | Structure-based evaluation of selective and non-selective small molecules that block HIV-1 TAT and PCAF association | | Descriptor: | (3E)-4-(1-METHYL-1H-INDOL-3-YL)BUT-3-EN-2-ONE, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Godbole, S, Muller, M, Yan, S, Sanchez, R, Zhou, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure-based evaluation of selective nad non-selective small molecules that block hiv-1 tat and pcaf association
TO BE PUBLISHED
|
|
1ZTR
 
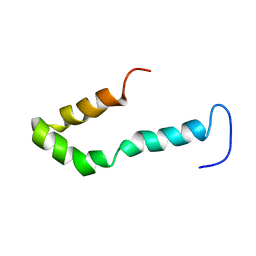 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
1ZTS
 
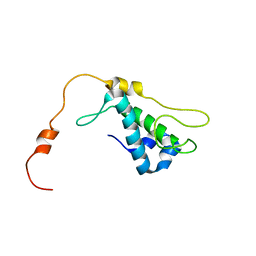 | | Solution Structure of Bacillus Subtilis Protein YQBG: Northeast Structural Genomics Consortium Target SR215 | | Descriptor: | Hypothetical protein yqbG | | Authors: | Liu, G, Shen, Y, Xiao, R, Acton, T, Ma, L, Joachimiak, A, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-27 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein yqbG from Bacillus subtilis reveals a novel alpha-helical protein fold.
Proteins, 62, 2006
|
|
1ZU1
 
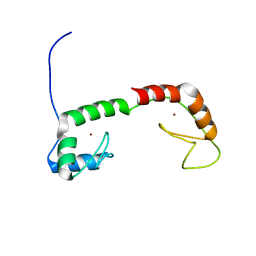 | | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa | | Descriptor: | RNA binding protein ZFa, ZINC ION | | Authors: | Moller, H.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-05-29 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc fingers of the Xenopus laevis double-stranded RNA-binding protein ZFa
J.Mol.Biol., 351, 2005
|
|
1ZU2
 
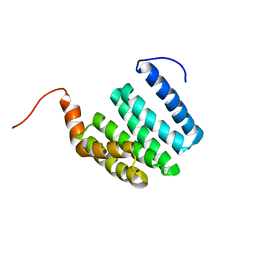 | |
1ZUB
 
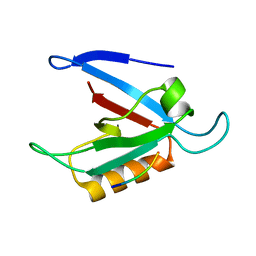 | | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide | | Descriptor: | ELKS1b, Regulating synaptic membrane exocytosis protein 1 | | Authors: | Lu, J, Li, H, Wang, Y, Sudhof, T.C, Rizo, J. | | Deposit date: | 2005-05-30 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RIM1alpha PDZ Domain in Complex with an ELKS1b C-terminal Peptide
J.Mol.Biol., 352, 2005
|
|
1ZUE
 
 | | Revised Solution Structure of DLP-2 | | Descriptor: | Defensin-like peptide 2/4 | | Authors: | Torres, A.M, Tsampazi, C, Geraghty, D.P, Bansal, P.S, Alewood, P.F, Kuchel, P.W. | | Deposit date: | 2005-05-30 | | Release date: | 2005-08-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | D-amino acid residue in a defensin-like peptide from platypus venom: effect on structure and chromatographic properties.
Biochem.J., 391, 2005
|
|
1ZUF
 
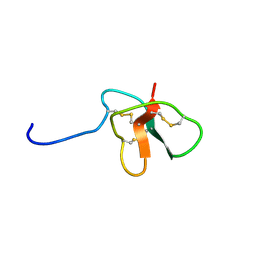 | | Solution Structure of DLP-4 | | Descriptor: | Defensin-like peptide 2/4 | | Authors: | Torres, A.M, Tsampazi, C, Geraghty, D.P, Bansal, P.S, Alewood, P.F, Kuchel, P.W. | | Deposit date: | 2005-05-31 | | Release date: | 2005-08-02 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | D-amino acid residue in a defensin-like peptide from platypus venom: effect on structure and chromatographic properties.
Biochem.J., 391, 2005
|
|
1ZUG
 
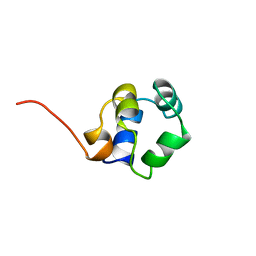 | | STRUCTURE OF PHAGE 434 CRO PROTEIN, NMR, 20 STRUCTURES | | Descriptor: | PHAGE 434 CRO PROTEIN | | Authors: | Padmanabhan, S, Jimenez, M.A, Gonzalez, C, Sanz, J.M, Gimenez-Gallego, G, Rico, M. | | Deposit date: | 1997-03-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure and stability of phage 434 Cro protein.
Biochemistry, 36, 1997
|
|
1ZUV
 
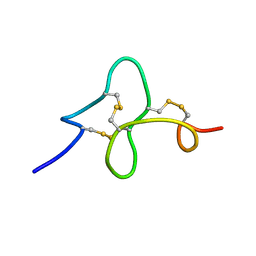 | | 24 NMR structures of AcAMP2-Like Peptide with Phenylalanine 18 mutated to Tryptophan | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-01 | | Release date: | 2005-12-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
1ZV6
 
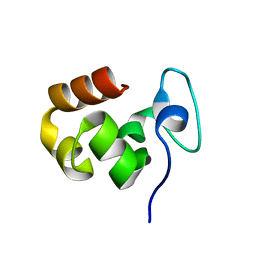 | |
1ZW7
 
 | | Elimination of the C-cap in Ubiquitin Structure, Dynamics and Thermodynamic Consequences | | Descriptor: | Ubiquitin | | Authors: | Ermolenko, D.N, Dangi, B, Gronenborn, A.M, Makhatadze, G.I. | | Deposit date: | 2005-06-03 | | Release date: | 2006-05-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Elimination of the C-cap in ubiquitin-structure, dynamics and thermodynamic consequences.
Biophys.Chem., 126, 2007
|
|
1ZW8
 
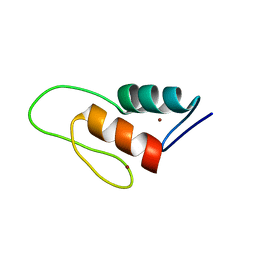 | | Solution structure of a ZAP1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae | | Descriptor: | ZINC ION, Zinc-responsive transcriptional regulator ZAP1 | | Authors: | Wang, Z, Feng, L.S, Venkataraman, K, Matskevich, V.A, Parasuram, P, Laity, J.H. | | Deposit date: | 2005-06-03 | | Release date: | 2006-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Zap1 zinc-responsive domain provides insights into metalloregulatory transcriptional repression in Saccharomyces cerevisiae.
J.Mol.Biol., 357, 2006
|
|
1ZWM
 
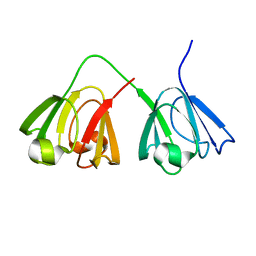 | | NMR structure of murine gamma-S crystallin | | Descriptor: | Gamma crystallin S | | Authors: | Wu, Z, Delaglio, F, Wyatt, K, Wistow, G, Bax, A. | | Deposit date: | 2005-06-03 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of (gamma)S-crystallin by molecular fragment replacement NMR.
Protein Sci., 14, 2005
|
|
1ZWO
 
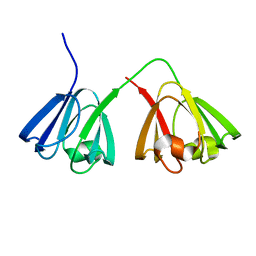 | | NMR structure of murine gamma-S crystallin | | Descriptor: | Gamma crystallin S | | Authors: | Wu, Z, Delaglio, F, Wyatt, K, Wistow, G, Bax, A. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of (gamma)S-crystallin by molecular fragment replacement NMR.
Protein Sci., 14, 2005
|
|
1ZWT
 
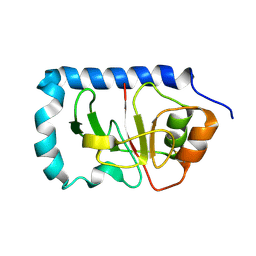 | | Structure of the globular head domain of the bundlin, BfpA, of the bundle-forming pilus of Enteropathogenic E.coli | | Descriptor: | Major structural subunit of bundle-forming pilus | | Authors: | Ramboarina, S, Fernandes, P.J, Daniell, S, Islam, S, Frankel, G, Booy, F, Donnenberg, M.S, Matthews, S. | | Deposit date: | 2005-06-06 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bundle-forming Pilus from Enteropathogenic Escherichia coli
J.Biol.Chem., 280, 2005
|
|
1ZWU
 
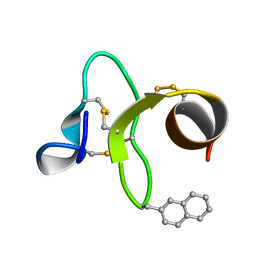 | | 30 NMR structures of AcAMP2-like peptide with non natural beta-(2-naphthyl)-alanine residue. | | Descriptor: | AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 (ACMP2) | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Freire, F, Aboitiz, N, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-06-06 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues.
Chemistry, 11, 2005
|
|
