8HTA
 
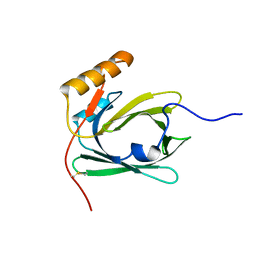 | |
8HVS
 
 | |
8HW9
 
 | | Solution structure of ubiquitin-like domain (UBL) of human ZFAND1 | | Descriptor: | AN1-type zinc finger protein 1 | | Authors: | Lai, C.H, Ko, K.T, Fan, P.J, Yu, T.A, Chang, C.F, Hsu, S.T.D. | | Deposit date: | 2022-12-29 | | Release date: | 2024-01-31 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the ZFAND1-p97 interaction involved in stress granule clearance.
J.Biol.Chem., 300, 2024
|
|
8HWU
 
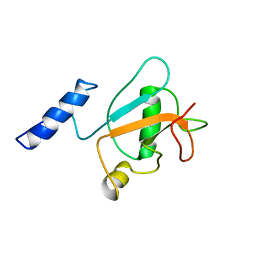
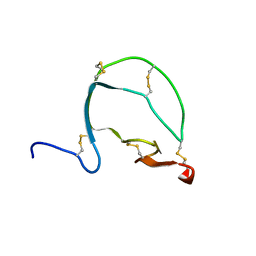 | | Solution structure of frog peptide LL-TIL | | Descriptor: | LL-TIL | | Authors: | Rami, M.J, Sarma, S.P. | | Deposit date: | 2023-01-03 | | Release date: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural, Functional, and Mutational Studies of a Potent Subtilisin Inhibitor from Budgett's Frog, Lepidobatrachus laevis.
Biochemistry, 62, 2023
|
|
8HZQ
 
 | | Bacillus subtilis SepF protein assembly (wild type) | | Descriptor: | Cell division protein SepF | | Authors: | Liu, W. | | Deposit date: | 2023-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular basis for curvature formation in SepF polymerization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8HZT
 
 | |
8HZW
 
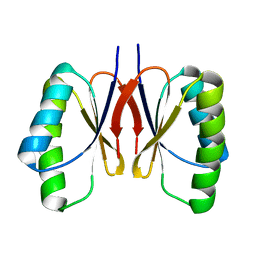
 | | The NMR structure of noursinH11W peptide | | Descriptor: | noursinH11W | | Authors: | Yao, H, Li, Y, Zhang, T, Gao, J, Wang, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Discovery and biosynthesis of tricyclic copper-binding ribosomal peptides containing histidine-to-butyrine crosslinks.
Nat Commun, 14, 2023
|
|
8I25
 
 | | NMR structure of Toxoplasma gondii PDCD5 (trans form) | | Descriptor: | Programmed cell death 5 protein | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Proline Isomerization and Molten Globular Property of TgPDCD5 Secreted from Toxoplasma gondii Confers Its Regulation of Heparin Sulfate Binding.
Jacs Au, 4, 2024
|
|
8I26
 
 | | NMR structure of Toxoplasma gondii PDCD5 (cis form) | | Descriptor: | Programmed cell death 5 protein | | Authors: | Lin, M.H, Hsu, C.H. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Proline Isomerization and Molten Globular Property of TgPDCD5 Secreted from Toxoplasma gondii Confers Its Regulation of Heparin Sulfate Binding.
Jacs Au, 4, 2024
|
|
8I43
 
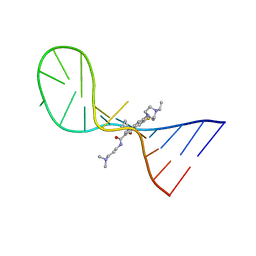 | |
8I44
 
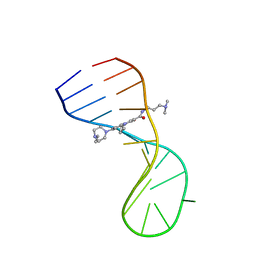 | |
8I45
 
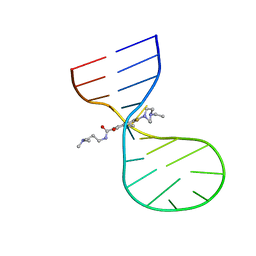 | |
8I46
 
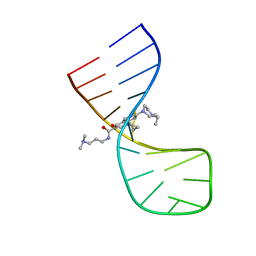 | |
8I53
 
 | |
8IB0
 
 | |
8IEX
 
 | | Solution structure of AtWRKY11-DBD | | Descriptor: | Probable WRKY transcription factor 11, ZINC ION | | Authors: | Dong, X, Hu, Y.F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of Arabidopsis transcription factor WRKY11.
Biochem.Biophys.Res.Commun., 653, 2023
|
|
8IF5
 
 | | AFB1-AF26 APTAMER COMPLEX | | Descriptor: | AFB1 DNA aptamer (26-MER), AFLATOXIN B1 | | Authors: | Xu, G.H, Wang, C, Li, C.G. | | Deposit date: | 2023-02-17 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer.
Nucleic Acids Res., 51, 2023
|
|
8IJC
 
 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8IKQ
 
 | |
8IL1
 
 | |
8IL2
 
 | |
8IL6
 
 | |
8IM5
 
 | |
8IMH
 
 | |
8IMQ
 
 | |
