7UGC
 
 | | Structure of the N-terminal domain of ViaA | | Descriptor: | Protein ViaA | | Authors: | Lemak, A, Reichheld, S, Bhandari, V, Houliston, S, Arrowsmith, C.H, Sharpe, S, Houry, W.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of ViaA
To Be Published
|
|
7UH9
 
 | | NMR structure of the cNTnC-cTnI chimera bound to W8 | | Descriptor: | CALCIUM ION, N-(7-aminoheptyl)-5-chloronaphthalene-1-sulfonamide, Troponin C, ... | | Authors: | Cai, F, Kampourakis, T, Cockburn, K.T, Sykes, B.D. | | Deposit date: | 2022-03-26 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Drugging the Sarcomere, a Delicate Balance: Position of N-Terminal Charge of the Inhibitor W7.
Acs Chem.Biol., 17, 2022
|
|
7UHA
 
 | | NMR structure of the cNTnC-cTnI chimera bound to W6 | | Descriptor: | CALCIUM ION, N-(5-aminopentyl)-5-chloronaphthalene-1-sulfonamide, Troponin C, ... | | Authors: | Cai, F, Kampourakis, T, Cockburn, K.T, Sykes, B.D. | | Deposit date: | 2022-03-26 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Drugging the Sarcomere, a Delicate Balance: Position of N-Terminal Charge of the Inhibitor W7.
Acs Chem.Biol., 17, 2022
|
|
7UI5
 
 | | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9 | | Descriptor: | CALCIUM ION, Protein S100-A9 | | Authors: | Reardon, P.N, Harman, J.L, Costello, S.M, Warren, G.D, Phillips, S.R, Connor, P.J, Marqusee, S, Harms, M.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UMC
 
 | |
7UMD
 
 | |
7UME
 
 | |
7UNX
 
 | | NMR solution structure of xanthusin-1 | | Descriptor: | Xanthusin-1 | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Discovery of Five Classes of Bacterial Defensins: Ancestral Precursors of Defensins from Eukarya?
Acs Omega, 2024
|
|
7UO6
 
 | |
7UQT
 
 | | Solution NMR structure of hexahistidine tagged QseM (6H-QseM) | | Descriptor: | Quorum sensing master protein | | Authors: | Hall, D.A, Solomon, P.D, Bond, C.S, Ramsay, J.P, Mackay, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | DUF2285 is a novel helix-turn-helix domain variant that orchestrates both activation and antiactivation of conjugative element transfer in proteobacteria.
Nucleic Acids Res., 51, 2023
|
|
7URJ
 
 | |
7UV1
 
 | | Vicilin Ana o 1.0101 leader sequence residues 20-75 | | Descriptor: | Vicilin-like protein | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV2
 
 | | Ana o 1 Leader Sequence Residues 82-132 | | Descriptor: | Vicilin-like protein | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV3
 
 | | Pis v 3.0101 Vicilin Leader Sequence Residues 5-52 | | Descriptor: | Vicilin Pis v 3.0101 | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UV4
 
 | | Pis v 3.0101 vicilin leader sequence residues 56-115 | | Descriptor: | Vicilin Pis v 3.0101 | | Authors: | Mueller, G.A, Foo, A.C.Y, DeRose, E.F. | | Deposit date: | 2022-04-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure and IgE Cross-Reactivity among Cashew, Pistachio, Walnut, and Peanut Vicilin-Buried Peptides.
J.Agric.Food Chem., 71, 2023
|
|
7UWY
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 29_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 29_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UWZ
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 33_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UZL
 
 | |
7V06
 
 | |
7V0V
 
 | | GFP Nanobody NMR Structure | | Descriptor: | Anti-GFP Nanobody | | Authors: | Mueller, G.A. | | Deposit date: | 2022-05-11 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nanobody Paratope Ensembles in Solution Characterized by MD Simulations and NMR.
Int J Mol Sci, 23, 2022
|
|
7V3T
 
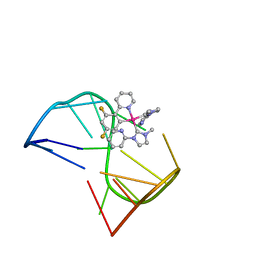 | | Solution structure of thrombin binding aptamer G-quadruplex bound a self-adaptive small molecule with rotated ligands | | Descriptor: | 11,13-bis(fluoranyl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-7$l^{4}-aza-8$l^{4}-platinatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene, TBA G4 DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Liu, W, Zhu, B.C, Mao, Z.W. | | Deposit date: | 2021-08-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound.
Chem Sci, 13, 2022
|
|
7V5E
 
 | |
7V5F
 
 | |
7V6V
 
 | |
7VB2
 
 | | Solution structure of human ribosomal protein uL11 | | Descriptor: | 60S ribosomal protein L12 | | Authors: | Lee, K.M, Wong, K.B. | | Deposit date: | 2021-08-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The flexible N-terminal motif of uL11 unique to eukaryotic ribosomes interacts with P-complex and facilitates protein translation.
Nucleic Acids Res., 50, 2022
|
|
