2N0O
 
 | |
2N0Q
 
 | | N2-dG-IQ modified DNA at the G1 position of the NarI recognition sequence | | Descriptor: | DNA_(5'-D(*CP*TP*CP*(IQG)P*GP*CP*GP*CP*CP*AP*TP*C)-3'), DNA_(5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3') | | Authors: | Stavros, K, Hawkins, E, Rizzo, C, Stone, M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Base-Displaced Intercalated Conformation of the 2-Amino-3-methylimidazo[4,5-f]quinoline N(2)-dG DNA Adduct Positioned at the Nonreiterated G(1) in the NarI Restriction Site.
Chem.Res.Toxicol., 28, 2015
|
|
2N0R
 
 | |
2N0S
 
 | | HADDOCK model of ferredoxin and [FeFe] hydrogenase complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Fe-hydrogenase, Ferredoxin, ... | | Authors: | Rumpel, S, Siebel, J, Fares, C, Reijerse, E, Lubitz, W. | | Deposit date: | 2015-03-13 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into the Complex of Ferredoxin and [FeFe] Hydrogenase from Chlamydomonas reinhardtii.
Chembiochem, 16, 2015
|
|
2N0T
 
 | | Structural ensemble of the enzyme cyclophilin reveals an orchestrated mode of action at atomic resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Chi, C.N, Voegeli, B, Bibow, S, Strotz, D, Orts, J, Guntert, P, Riek, R. | | Deposit date: | 2015-03-13 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Structural Ensemble for the Enzyme Cyclophilin Reveals an Orchestrated Mode of Action at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2N0U
 
 | | Mdmx-057 | | Descriptor: | 4-[(4S,5R)-4-(3-chlorophenyl)-5-(4-chlorophenyl)-1-(3-oxidanylidenepiperazin-1-yl)carbonyl-4,5-dihydroimidazol-2-yl]-3-propan-2-yloxy-benzenecarbonitrile, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2N0V
 
 | | Backbone 1H, Chemical Shift Assignments for Cn-APM1 | | Descriptor: | Antimicrobial peptide 1 | | Authors: | Santana, M.J, Oliveira, A.L, Queiroz Jr, L.K, Mandal, S.M, Matos, C.O, Dias, R.O, Franco, O.L, Liao, L.M. | | Deposit date: | 2015-03-17 | | Release date: | 2015-12-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural insights into Cn-AMP1, a short disulfide-free multifunctional peptide from green coconut water.
Febs Lett., 589, 2015
|
|
2N0W
 
 | | Mdmx-SJ212 | | Descriptor: | 4-({(4S,5R)-4-(5-bromo-2-fluorophenyl)-5-(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2N0X
 
 | | Three dimensional structure of EPI-X4, a human albumin-derived peptide that regulates innate immunity through the CXCR4/CXCL12 chemotactic axis and antagonizes HIV-1 entry | | Descriptor: | Serum albumin | | Authors: | Perez-Castells, J, Canales, A, Jimenez-Barbero, J, Gimenez-Gallego, G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Discovery and characterization of an endogenous CXCR4 antagonist.
Cell Rep, 11, 2015
|
|
2N0Y
 
 | | NMR structure of the complex between the C-terminal domain of the Rift Valley fever virus protein NSs and the PH domain of the Tfb1 subunit of TFIIH | | Descriptor: | Non-structural protein NS-S, RNA polymerase II transcription factor B subunit 1 | | Authors: | Cyr, N, de la Fuente, C, Lecoq, L, Guendel, I, Chabot, P.R, Kehn-Hall, K, Omichinski, J.G. | | Deposit date: | 2015-03-18 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Omega XaV motif in the Rift Valley fever virus NSs protein is essential for degrading p62, forming nuclear filaments and virulence.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2N0Z
 
 | |
2N10
 
 | |
2N11
 
 | |
2N12
 
 | |
2N13
 
 | |
2N14
 
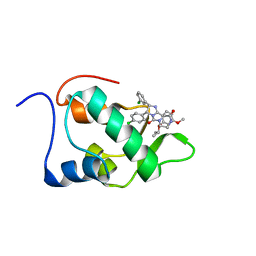 | | Mdmx-295 | | Descriptor: | 4-({(4S,5R)-4-(3-chlorophenyl)-5-(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-20 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2N16
 
 | |
2N17
 
 | | NMR structure of a Kazal-type serine protease inhibitor from the subterranean termite defense gland of Coptotermes formosanus Shiraki soldiers | | Descriptor: | Lysozyme-Protease Inhibitor Protein | | Authors: | Negulescu, H, Guo, Y, Garner, T.P, Goodwin, O.Y, Henderson, G, Laine, R.A, Macnaughtan, M.A. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Kazal-Type Serine Protease Inhibitor from the Defense Gland Secretion of the Subterranean Termite Coptotermes formosanus Shiraki.
Plos One, 10, 2015
|
|
2N18
 
 | |
2N19
 
 | | STIL binding to the Polo-box domain 3 of PLK4 regulates centriole duplication | | Descriptor: | Serine/threonine-protein kinase PLK4 | | Authors: | Boehm, R, Arquint, C, Gabryjonczyk, A, Imseng, S, Sauer, E, Hiller, S, Nigg, E, Maier, T. | | Deposit date: | 2015-03-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | STIL binding to Polo-box 3 of PLK4 regulates centriole duplication.
Elife, 4, 2015
|
|
2N1A
 
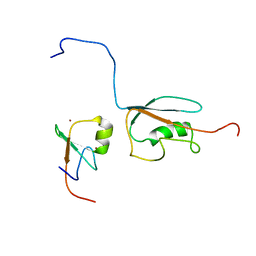 | | Docked structure between SUMO1 and ZZ-domain from CBP | | Descriptor: | CREB-binding protein, Small ubiquitin-related modifier 1, ZINC ION | | Authors: | Diehl, C. | | Deposit date: | 2015-03-26 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Analysis of a Complex between Small Ubiquitin-like Modifier 1 (SUMO1) and the ZZ Domain of CREB-binding Protein (CBP/p300) Reveals a New Interaction Surface on SUMO.
J.Biol.Chem., 291, 2016
|
|
2N1B
 
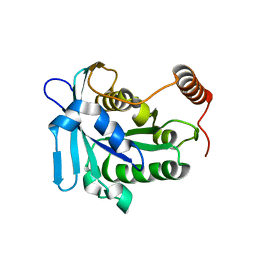 | |
2N1C
 
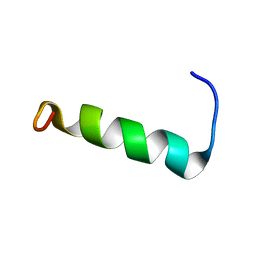 | | Structure of PvHCt, an antimicrobial peptide from shrimp litopenaeus vannamei | | Descriptor: | Hemocyanin subunit L2 | | Authors: | Petit, V.W, Rolland, J.L, Blond, A, Djediat, C, Peduzzi, J, Goulard, C, Bachere, E, Dupont, J, Destoumieux-Garzon, D, Rebuffat, S. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A hemocyanin-derived antimicrobial peptide from the penaeid shrimp adopts an alpha-helical structure that specifically permeabilizes fungal membranes.
Biochim.Biophys.Acta, 1860, 2015
|
|
2N1D
 
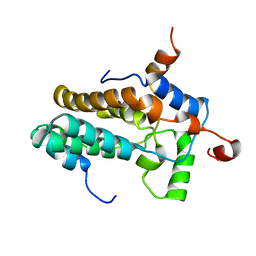 | | Solution structure of the MRG15-MRGBP complex | | Descriptor: | MRG/MORF4L-binding protein, Mortality factor 4-like protein 1 | | Authors: | Xie, T, Zmysloski, A.M, Zhang, Y, Radhakrishnan, I. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Multi-specificity of MRG Domains.
Structure, 23, 2015
|
|
2N1E
 
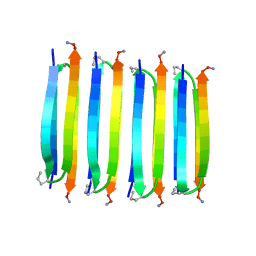 | | MAX1 peptide fibril | | Descriptor: | MAX1 peptide | | Authors: | Nagy-Smith, K, Moore, E, Schneider, J, Tycko, R. | | Deposit date: | 2015-03-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-30 | | Method: | SOLID-STATE NMR | | Cite: | Molecular structure of monomorphic peptide fibrils within a kinetically trapped hydrogel network.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
