1ZBN
 
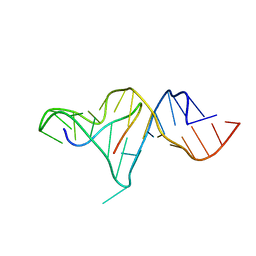 | |
1ZC1
 
 | | Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites | | Descriptor: | Ubiquitin fusion degradation protein 1 | | Authors: | Park, S, Isaacson, R, Kim, H.T, Silver, P.A, Wagner, G. | | Deposit date: | 2005-04-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ufd1 Exhibits the AAA-ATPase Fold with Two Distinct Ubiquitin Interaction Sites
Structure, 13, 2005
|
|
1ZDV
 
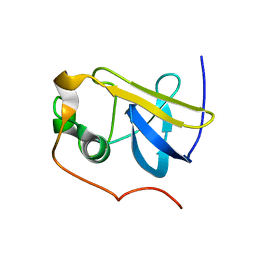 | | Solution Structure of the type 1 pilus assembly platform FimD(25-139) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZDX
 
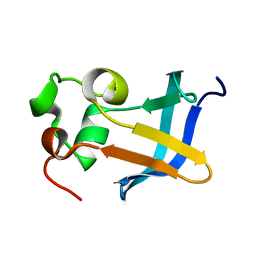 | | Solution Structure of the type 1 pilus assembly platform FimD(25-125) | | Descriptor: | Outer membrane usher protein fimD | | Authors: | Nishiyama, M, Horst, R, Herrmann, T, Vetsch, M, Bettendorff, P, Ignatov, O, Grutter, M, Wuthrich, K, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-04-15 | | Release date: | 2005-06-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD.
Embo J., 24, 2005
|
|
1ZE7
 
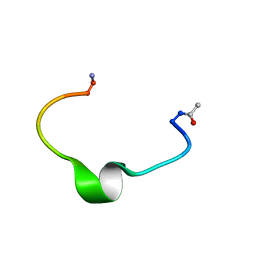 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide in water solution at pH 6.5 | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2013-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
1ZE9
 
 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide complexed with a zinc (II) cation | | Descriptor: | 16-mer from Alzheimer's disease amyloid Protein, ZINC ION | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-05-03 | | Last modified: | 2013-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural changes of region 1-16 of the Alzheimer disease amyloid beta-peptide upon zinc binding and in vitro aging.
J.Biol.Chem., 281, 2006
|
|
1ZEC
 
 | | NMR Solution structure of NEF1-25, 20 structures | | Descriptor: | NEF1-25 | | Authors: | Barnham, K.J, Monks, S.A, Hinds, M.G, Azad, A.A, Norton, R.S. | | Deposit date: | 1996-12-18 | | Release date: | 1998-01-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a polypeptide from the N terminus of the HIV protein Nef.
Biochemistry, 36, 1997
|
|
1ZFD
 
 | | SWI5 ZINC FINGER DOMAIN 2, NMR, 45 STRUCTURES | | Descriptor: | SWI5, ZINC ION | | Authors: | Neuhaus, D, Nakaseko, Y, Schwabe, J.W.R, Rhodes, D, Klug, A. | | Deposit date: | 1996-04-04 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of beta-sheet.
J.Mol.Biol., 228, 1992
|
|
1ZFO
 
 | | AMINO-TERMINAL LIM-DOMAIN PEPTIDE OF LASP-1, NMR | | Descriptor: | LASP-1, ZINC ION | | Authors: | Hammarstrom, A, Berndt, K.D, Sillard, R, Adermann, K, Otting, G. | | Deposit date: | 1996-05-06 | | Release date: | 1996-11-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a naturally-occurring zinc-peptide complex demonstrates that the N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit.
Biochemistry, 35, 1996
|
|
1ZFS
 
 | |
1ZFU
 
 | | Plectasin:A peptide antibiotic with therapeutic potential from a saprophytic fungus | | Descriptor: | Plectasin | | Authors: | Mygind, P.H, Fischer, R.L, Schnorr, K, Hansen, M.T, Sonksen, C.P, Ludvigsen, S, Raventos, D, Buskov, S, Christensen, B, De Maria, L, Taboureau, O, Yaver, D, Elvig-Jorgensen, S.G, Sorensen, M.V, Christensen, B.E, Kjaerulf, S, Frimodt-Moller, N, Lehrer, R.I, Zasloff, M, Kristensen, H.H. | | Deposit date: | 2005-04-20 | | Release date: | 2005-10-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus.
Nature, 437, 2005
|
|
1ZG2
 
 | | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2. | | Descriptor: | Hypothetical UPF0213 protein BH0048 | | Authors: | Aramini, J.M, Swapna, G.V.T, Xiao, R, Ma, L, Shastry, R, Ciano, M, Acton, T.B, Liu, J, Rost, B, Cort, J.R, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-20 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the UPF0213 protein BH0048 from Bacillus halodurans. Northeast Structural Genomics target BhR2.
To be Published
|
|
1ZGG
 
 | |
1ZGU
 
 | | Solution structure of the human Mms2-Ubiquitin complex | | Descriptor: | Ubiquitin, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Lewis, M.J, Saltibus, L.F, Hau, D.D, Xiao, W, Spyracopoulos, L. | | Deposit date: | 2005-04-22 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Non-Covalent Interaction Between Ubiquitin and the Ubiquitin Conjugating Enzyme Variant Human MMS2.
J.Biomol.Nmr, 34, 2006
|
|
1ZGW
 
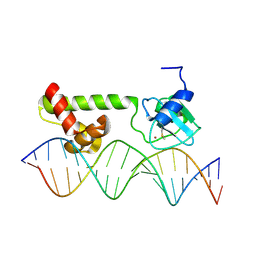 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1ZHC
 
 | |
1ZIT
 
 | |
1ZJQ
 
 | |
1ZK6
 
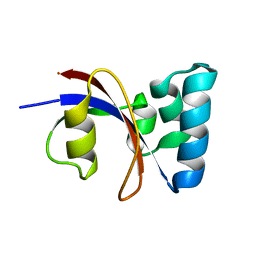 | | NMR solution structure of B. subtilis PrsA PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Tossavainen, H, Permi, P, Purhonen, S.L, Sarvas, M, Kilpelainen, I, Seppala, R. | | Deposit date: | 2005-05-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis
Febs Lett., 580, 2006
|
|
1ZKH
 
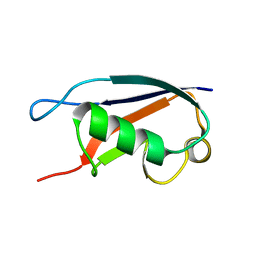 | | Solution structure of a human ubiquitin-like domain in SF3A1 | | Descriptor: | Splicing factor 3 subunit 1 | | Authors: | Lukin, J.A, Dhe-Paganon, S, Guido, V, Lemak, A, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human ubiquitin-like domain in SF3A1
To be Published
|
|
1ZLL
 
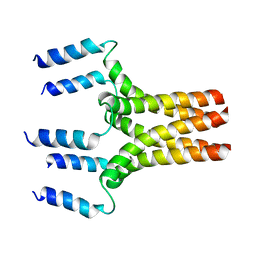 | |
1ZMZ
 
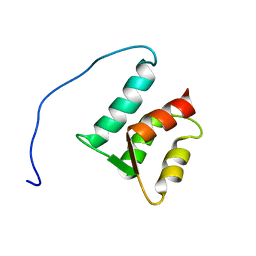 | | Solution structure of the N-terminal domain (M1-S98) of human centrin 2 | | Descriptor: | Centrin-2 | | Authors: | Yang, A, Miron, S, Duchambon, P, Assairi, L, Blouquit, Y, Craescu, C.T. | | Deposit date: | 2005-05-11 | | Release date: | 2006-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly
Biochemistry, 45, 2006
|
|
1ZNT
 
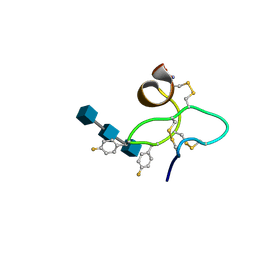 | | 18 NMR structures of AcAMP2-Like Peptide with non Natural Fluoroaromatic Residue (AcAMP2F18Pff/Y20Pff) complex with N,N,N-triacetylchitotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMARANTHUS CAUDATUS ANTIMICROBIAL PEPTIDE 2 | | Authors: | Chavez, M.I, Andreu, C, Vidal, P, Aboitiz, N, Freire, F, Groves, P, Asensio, J.L, Asensio, G, Muraki, M, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2005-05-12 | | Release date: | 2005-12-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | On the Importance of Carbohydrate-Aromatic Interactions for the Molecular Recognition of Oligosaccharides by Proteins: NMR Studies of the Structure and Binding Affinity of AcAMP2-like Peptides with Non-Natural Naphthyl and Fluoroaromatic Residues
Chemistry, 11, 2005
|
|
1ZNU
 
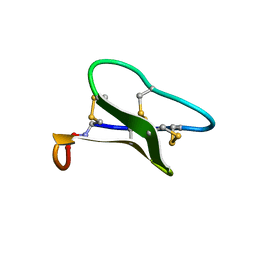 | | Structure of cyclotide Kalata B1 in DPC micelles solution | | Descriptor: | Kalata B1 | | Authors: | Shenkarev, Z.O, Nadezhdin, K.D, Sobol, V.A, Sobol, A.G, Skjeldal, L, Arseniev, A.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-04-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle.
Febs J., 273, 2006
|
|
1ZO0
 
 | |
