1RJT
 
 | |
1RJV
 
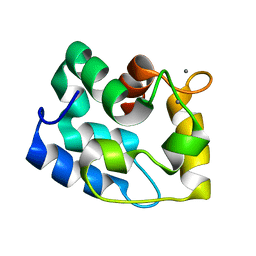 | | Solution Structure of Human alpha-Parvalbumin refined with a paramagnetism-based strategy | | Descriptor: | CALCIUM ION, Parvalbumin alpha | | Authors: | Baig, I, Bertini, I, Del Bianco, C, Gupta, Y.K, Lee, Y.M, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-11-20 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Paramagnetism-Based Refinement Strategy for the Solution Structure of Human alpha-Parvalbumin.
Biochemistry, 43, 2004
|
|
1RK7
 
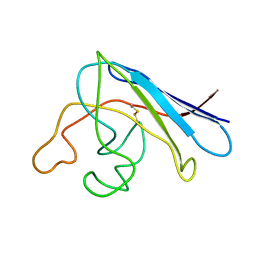 | | Solution structure of apo Cu,Zn Superoxide Dismutase: role of metal ions in protein folding | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Banci, L, Bertini, I, Cramaro, F, Del Conte, R, Viezzoli, M.S. | | Deposit date: | 2003-11-21 | | Release date: | 2003-12-02 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Apo Cu,Zn Superoxide Dismutase: Role of Metal Ions in Protein Folding
Biochemistry, 42, 2003
|
|
1RKK
 
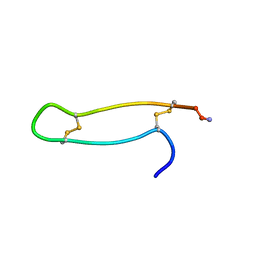 | |
1RKL
 
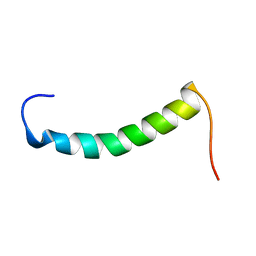 | |
1RKN
 
 | | Solution structure of 1-110 fragment of Staphylococcal Nuclease with G88W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D.S, Feng, Y.G, Ye, K.Q, Shan, L, Wang, J.F. | | Deposit date: | 2003-11-22 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
1RL1
 
 | | Solution structure of human Sgt1 CS domain | | Descriptor: | Suppressor of G2 allele of SKP1 homolog | | Authors: | Lee, Y.-T, Jacob, J, Michowski, W, Nowotny, M, Kuznicki, J, Chazin, W.J. | | Deposit date: | 2003-11-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Human Sgt1 Binds HSP90 through the CHORD-Sgt1 Domain and Not the Tetratricopeptide Repeat Domain
J.Biol.Chem., 279, 2004
|
|
1RL5
 
 | | NMR structure with tightly bound water molecule of cytotoxin I from Naja oxiana in aqueous solution (major form) | | Descriptor: | Cytotoxin 1 | | Authors: | Dubinnyi, M.A, Pustovalova, Y.E, Dubovskii, P.V, Utkin, Y.N, Arseniev, A.S. | | Deposit date: | 2003-11-25 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Interaction of three-finger toxins with phospholipid membranes: comparison of S- and P-type cytotoxins.
Biochem.J., 387, 2005
|
|
1RLP
 
 | | TWO BINDING ORIENTATIONS FOR PEPTIDES TO SRC SH3 DOMAIN: DEVELOPMENT OF A GENERAL MODEL FOR SH3-LIGAND INTERACTIONS | | Descriptor: | C-SRC TYROSINE KINASE SH3 DOMAIN, PROLINE-RICH LIGAND RLP2 (RALPPLPRY) | | Authors: | Feng, S, Chen, J.K, Yu, H, Simon, J.A, Schreiber, S.L. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Two binding orientations for peptides to the Src SH3 domain: development of a general model for SH3-ligand interactions.
Science, 266, 1994
|
|
1RMK
 
 | | Solution structure of conotoxin MrVIB | | Descriptor: | Mu-O-conotoxin MrVIB | | Authors: | Daly, N.L, Ekberg, J.A, Thomas, L, Adams, D.J, Lewis, R.J, Craik, D.J. | | Deposit date: | 2003-11-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structures of muO-conotoxins from Conus marmoreus. Inhibitors of tetrodotoxin (TTX)-sensitive and TTX-resistant sodium channels in mammalian sensory neurons
J.Biol.Chem., 279, 2004
|
|
1RMX
 
 | | A SHORT LEXITROPSIN THAT RECOGNIZES THE DNA MINOR GROOVE AT 5'-ACTAGT-3': UNDERSTANDING THE ROLE OF ISOPROPYL-THIAZOLE | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3', N-[3-(DIMETHYLAMINO)PROPYL]-2-({[4-({[4-(FORMYLAMINO)-1-METHYL-1H-PYRROL-2-YL]CARBONYL}AMINO)-1-METHYL-1H-PYRROL-2-YL]CARBONYL}AMINO)-5-ISOPROPYL-1,3-THIAZOLE-4-CARBOXAMIDE | | Authors: | Anthony, N.G, Johnston, B.F, Khalaf, A.I, Mackay, S.P, Parkinson, J.A, Suckling, C.J, Waigh, R.D. | | Deposit date: | 2003-11-28 | | Release date: | 2004-08-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Short Lexitropsin that Recognizes the DNA Minor Groove at 5'-ACTAGT-3': Understanding the Role of Isopropyl-thiazole.
J.Am.Chem.Soc., 126, 2004
|
|
1RN9
 
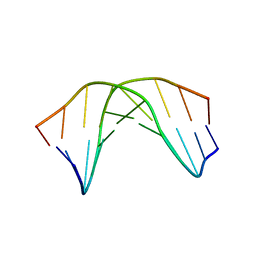 | | A SHORT LEXITROPSIN THAT RECOGNIZES THE DNA MINOR GROOVE AT 5'-ACTAGT-3': UNDERSTANDING THE ROLE OF ISOPROPYL-THIAZOLE | | Descriptor: | 5'-D(*CP*GP*AP*CP*TP*AP*GP*TP*CP*G)-3' | | Authors: | Anthony, N.G, Johnston, B.F, Khalaf, A.I, Mackay, S.P, Parkinson, J.A, Suckling, C.J, Waigh, R.D. | | Deposit date: | 2003-12-01 | | Release date: | 2004-08-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Short Lexitropsin that Recognizes the DNA Minor Groove at 5'-ACTAGT-3': Understanding the Role of Isopropyl-thiazole.
J.Am.Chem.Soc., 126, 2004
|
|
1RNK
 
 | |
1RO3
 
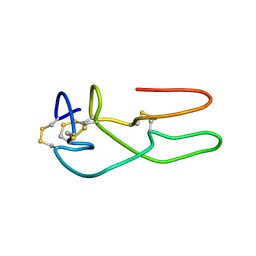 | | New structural insights on short disintegrin echistatin by NMR | | Descriptor: | Disintegrin echistatin | | Authors: | Monleon, D, Esteve, V, Calvete, J.J, Marcinkiewicz, C, Celda, B. | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Conformation and concerted dynamics of the integrin-binding site and the C-terminal region of echistatin revealed by homonuclear NMR
Biochem.J., 387, 2005
|
|
1RON
 
 | |
1ROO
 
 | | NMR SOLUTION STRUCTURE OF SHK TOXIN, NMR, 20 STRUCTURES | | Descriptor: | SHK TOXIN | | Authors: | Tudor, J.E, Pallaghy, P.K, Pennington, M.W, Norton, R.S. | | Deposit date: | 1996-01-11 | | Release date: | 1997-01-27 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ShK toxin, a novel potassium channel inhibitor from a sea anemone.
Nat.Struct.Biol., 3, 1996
|
|
1ROT
 
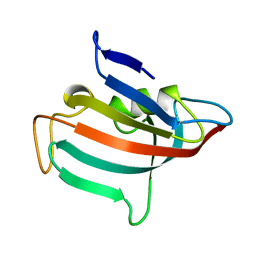 | | STRUCTURE OF FKBP59-I, THE N-TERMINAL DOMAIN OF A 59 KDA FK506-BINDING PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FKBP59-I | | Authors: | Craescu, C.T, Rouviere, N, Popescu, A, Cerpolini, E, Lebeau, M.-C, Baulieu, E.-E, Mispelter, J. | | Deposit date: | 1996-06-14 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the immunophilin-like domain of FKBP59 in solution.
Biochemistry, 35, 1996
|
|
1RPR
 
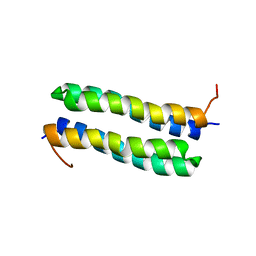 | | THE STRUCTURE OF COLE1 ROP IN SOLUTION | | Descriptor: | ROP | | Authors: | Eberle, W, Pastore, A, Klaus, W, Sander, C, Roesch, P. | | Deposit date: | 1991-10-09 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of ColE1 rop in solution.
J.Biomol.NMR, 1, 1991
|
|
1RPV
 
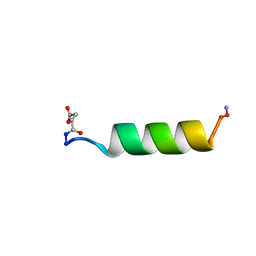 | | HIV-1 REV PROTEIN (RESIDUES 34-50) | | Descriptor: | HIV-1 REV PROTEIN | | Authors: | Scanlon, M.J, Fairlie, D.P, Craik, D.J, Englebretsen, D.R, West, M.L. | | Deposit date: | 1995-05-04 | | Release date: | 1995-10-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the RNA-binding peptide from human immunodeficiency virus (type 1) Rev.
Biochemistry, 34, 1995
|
|
1RQ6
 
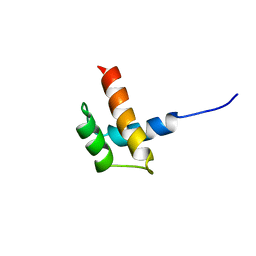 | | Solution structure of ribosomal protein S17E from Methanobacterium Thermoautotrophicum, Northeast Structural Genomics Consortium Target TT802 / Ontario Center for Structural Proteomics Target Mth0803 | | Descriptor: | 30S ribosomal protein S17e | | Authors: | Wu, B, Yee, A, Huang, Y.J, Ramelot, T.A, Semesi, A, Jung, J.W, Edward, A, Lee, W, Kennedy, M.A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-04 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of ribosomal protein S17E from Methanobacterium thermoautotrophicum: a structural homolog of the FF domain.
Protein Sci., 17, 2008
|
|
1RQ8
 
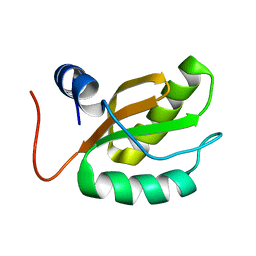 | |
1RQM
 
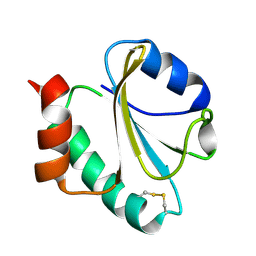 | | SOLUTION STRUCTURE OF THE K18G/R82E ALICYCLOBACILLUS ACIDOCALDARIUS THIOREDOXIN MUTANT | | Descriptor: | Thioredoxin | | Authors: | Leone, M, Di Lello, P, Ohlenschlager, O, Pedone, E.M, Bartolucci, S, Rossi, M, Di Blasio, B, Pedone, C, Saviano, M, Isernia, C, Fattorusso, R. | | Deposit date: | 2003-12-05 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the K18G/R82E Alicyclobacillus acidocaldarius Thioredoxin Mutant: A Molecular Analysis of Its Reduced Thermal Stability.
Biochemistry, 43, 2004
|
|
1RQS
 
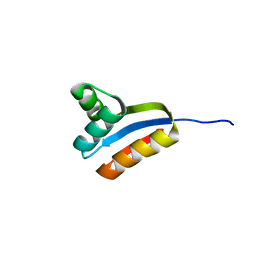 | | NMR structure of C-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQT
 
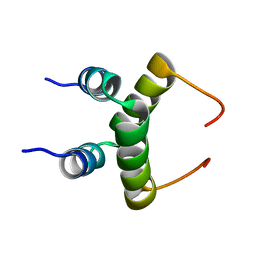 | | NMR structure of dimeric N-terminal domain of ribosomal protein L7 from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
1RQU
 
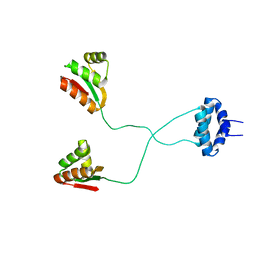 | | NMR structure of L7 dimer from E.coli | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome.
J.Biol.Chem., 279, 2004
|
|
