5OV2
 
 | |
5OVM
 
 | | Solution structure of lipase binding domain LID1 of foldase from Pseudomonas aeruginosa | | Descriptor: | Lipase chaperone | | Authors: | Viegas, A, Jaeger, K.-E, Etzkorn, M, Gohlke, H, Verma, N, Dollinger, P, Kovacic, F. | | Deposit date: | 2017-08-29 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic insights revealing how lipase binding domain MD1 of Pseudomonas aeruginosa foldase affects lipase activation.
Sci Rep, 10, 2020
|
|
5OWI
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 1) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
5OWJ
 
 | | The dynamic dimer structure of the chaperone Trigger Factor (conformer 2) | | Descriptor: | Trigger factor | | Authors: | Morgado, L, Burmann, B.M, Sharpe, T, Mazur, A, Hiller, S. | | Deposit date: | 2017-09-01 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dynamic dimer structure of the chaperone Trigger Factor.
Nat Commun, 8, 2017
|
|
5SXY
 
 | |
5SYQ
 
 | | Solution structure of Aquifex aeolicus Aq1974 | | Descriptor: | Uncharacterized protein aq_1974 | | Authors: | Sachleben, J.R, Gawlak, G, Hoey, R.J, Liu, G, Joachimiak, A, Montelione, G.T, Koide, S, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Aromatic claw: A new fold with high aromatic content that evades structural prediction.
Protein Sci., 26, 2017
|
|
5SZW
 
 | | NMR solution structure of the RRM1 domain of the post-transcriptional regulator HuR | | Descriptor: | ELAV-like protein 1 | | Authors: | Lixa, C, Mujo, A, Jendiroba, K.A, Almeida, F.C.L, Lima, L.M.T.R, Pinheiro, A.S. | | Deposit date: | 2016-08-15 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oligomeric transition and dynamics of RNA binding by the HuR RRM1 domain in solution.
J. Biomol. NMR, 72, 2018
|
|
5T0X
 
 | |
5T17
 
 | |
5T1N
 
 | |
5T1O
 
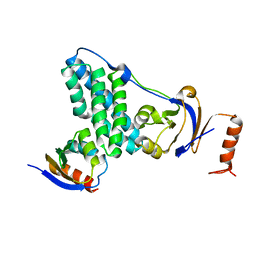 | | Solution-state NMR and SAXS structural ensemble of NPr (1-85) in complex with EIN-Ntr (170-424) | | Descriptor: | Phosphocarrier protein NPr, Phosphoenolpyruvate-protein phosphotransferase PtsP | | Authors: | Strickland, M, Stanley, A.M, Wang, G, Schwieters, C.D, Buchanan, S, Peterkofsky, A, Tjandra, N. | | Deposit date: | 2016-08-19 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure of the NPr:EIN(Ntr) Complex: Mechanism for Specificity in Paralogous Phosphotransferase Systems.
Structure, 24, 2016
|
|
5T3M
 
 | |
5T3Y
 
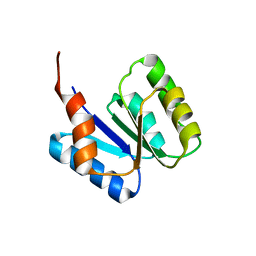 | |
5T42
 
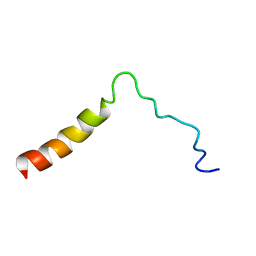 | | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity | | Descriptor: | Envelope glycoprotein | | Authors: | Lee, J, Nyenhuis, D.A, Nelson, E.A, Cafiso, D.S, White, J.M, Tamm, L.K. | | Deposit date: | 2016-08-28 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
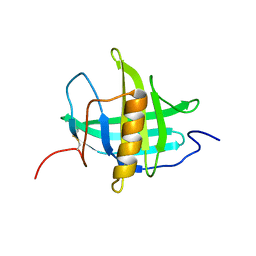
5T43
 
 | |
5T4R
 
 | |
5T56
 
 | |
5T7C
 
 | |
5T7Q
 
 | | TIRAP phosphoinositide-binding motif | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Capelluto, D.G.S, Ellena, J.F, Armstrong, G, Zhao, X, Xiao, S. | | Deposit date: | 2016-09-05 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Membrane targeting of TIRAP is negatively regulated by phosphorylation in its phosphoinositide-binding motif.
Sci Rep, 7, 2017
|
|
5T82
 
 | |
5T8A
 
 | | Recombinant cytotoxin-I from the venom of cobra N. oxiana | | Descriptor: | Cytotoxin 1 | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
5TBG
 
 | |
5TBN
 
 | |
5TBQ
 
 | |
5TBR
 
 | |
