5L6R
 
 | | PrP226* - Solution-state NMR structure of truncated human prion protein | | Descriptor: | Major prion protein | | Authors: | Kovac, V, Zupancic, B, Ilc, G, Curin Serbec, V, Plavec, J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Truncated prion protein PrP226* - A structural view on its role in amyloid disease.
Biochem. Biophys. Res. Commun., 484, 2017
|
|
5L7B
 
 | | Solution structure of the human SNF5/INI1 domain | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Allen, M.D, Zinzalla, G, Bycroft, M. | | Deposit date: | 2016-06-02 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex.
FEBS J., 285, 2018
|
|
5L7M
 
 | | Murin CXCL13 solution structure | | Descriptor: | C-X-C motif chemokine 13 | | Authors: | Monneau, Y.R, Lortat-Jacob, H. | | Deposit date: | 2016-06-03 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of CXCL13 and heparan sulfate binding show that GAG binding site and cellular signalling rely on distinct domains.
Open Biol, 7, 2017
|
|
5L82
 
 | |
5L85
 
 | |
5LAH
 
 | | NMR structure of the sea anemone peptide tau-AnmTx Ueq 12-1 with an uncommon fold | | Descriptor: | tau-AnmTx Ueq 12-1 | | Authors: | Mineev, K.S, Arseniev, A.S, Andreev, Y.A, Kozlov, S.A, Logashina, Y.A. | | Deposit date: | 2016-06-14 | | Release date: | 2017-05-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | New Disulfide-Stabilized Fold Provides Sea Anemone Peptide to Exhibit Both Antimicrobial and TRPA1 Potentiating Properties
Toxins, 9, 2017
|
|
5LAM
 
 | | Refined 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LAO
 
 | | S-nitrosylated 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Maslennikov, I, Kwiatkowski, W, Choe, S, Lipton, S.A, Guntert, P, Riek, R. | | Deposit date: | 2016-06-14 | | Release date: | 2016-08-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
5LBJ
 
 | |
5LCB
 
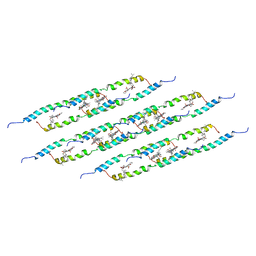 | | In situ atomic-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum obtained combining solid-state NMR spectroscopy, cryo electron microscopy and polarization spectroscopy | | Descriptor: | BACTERIOCHLOROPHYLL A, Bacteriochlorophyll c-binding protein | | Authors: | Nielsen, J.T, Kulminskaya, N.V, Bjerring, M, Linnanto, J.M, Ratsep, M, Pedersen, M, Lambrev, P.H, Dorogi, M, Garab, G, Thomsen, K, Jegerschold, C, Frigaard, N.U, Lindahl, M, Nielsen, N.C. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (26.5 Å), SOLID-STATE NMR | | Cite: | In situ high-resolution structure of the baseplate antenna complex in Chlorobaculum tepidum.
Nat Commun, 7, 2016
|
|
5LCI
 
 | |
5LCS
 
 | |
5LDL
 
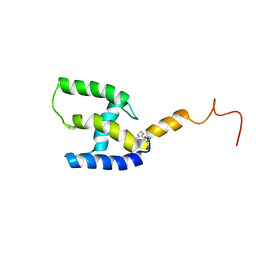 | | Myristoylated T41I/T78I mutant of M-PMV matrix protein | | Descriptor: | MYRISTIC ACID, myristoylated M-PMV matrix protein mutant | | Authors: | Kroupa, T, Hrabal, R. | | Deposit date: | 2016-06-27 | | Release date: | 2016-07-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Membrane Interactions of the Mason-Pfizer Monkey Virus Matrix Protein and Its Budding Deficient Mutants.
J.Mol.Biol., 428, 2016
|
|
5LFF
 
 | | NMR structure of peptide 2 targeting CXCR4 | | Descriptor: | ARG-ALA-CYS-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
5LFH
 
 | | NMR structure of peptide 10 targeting CXCR4 | | Descriptor: | ACE-ARG-ALA-DCY-ARG-PHE-PHE-CYS | | Authors: | Di Maro, S, Trotta, A.M, Brancaccio, D, Di Leva, F.S, La Pietra, V, Ierano, C, Napolitano, M, Portella, L, D'Alterio, C, Siciliano, R.A, Sementa, D, Tomassi, S, Carotenuto, A, Novellino, E, Scala, S, Marinelli, L. | | Deposit date: | 2016-07-01 | | Release date: | 2016-09-07 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Exploring the N-Terminal Region of C-X-C Motif Chemokine 12 (CXCL12): Identification of Plasma-Stable Cyclic Peptides As Novel, Potent C-X-C Chemokine Receptor Type 4 (CXCR4) Antagonists.
J.Med.Chem., 59, 2016
|
|
5LFI
 
 | | lactococcin A immunity protein | | Descriptor: | Lactococcin-A immunity protein | | Authors: | Persson, C, Fuochi, V, Pedersen, A, Karlsson, B.G, Nissen-Meyer, J, Kristiansen, P.E, Oppegard, C. | | Deposit date: | 2016-07-01 | | Release date: | 2016-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structure and Mutational Analysis of the Lactococcin A Immunity Protein.
Biochemistry, 55, 2016
|
|
5LFY
 
 | |
5LG0
 
 | | Solution NMR structure of Tryptophan to Alanine mutant of Arkadia RING domain. | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-05 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
5LG7
 
 | | Solution NMR structure of Tryptophan to Arginine mutant of Arkadia RING domain | | Descriptor: | E3 ubiquitin-protein ligase Arkadia, ZINC ION | | Authors: | Birkou, M, Chasapis, C.T, Loutsidou, A.K, Bentrop, D, Lelli, M, Herrmann, T, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Residue Specific Insight into the Arkadia E3 Ubiquitin Ligase Activity and Conformational Plasticity.
J. Mol. Biol., 429, 2017
|
|
5LG9
 
 | |
5LGF
 
 | |
5LGM
 
 | | Gp5.7 mutant L42A | | Descriptor: | Fusion protein 5.5/5.7 | | Authors: | Liu, B, Matthews, S. | | Deposit date: | 2016-07-07 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Gp5.7 mutant L42A
to be published
|
|
5LIG
 
 | | G-Quadruplex formed at the 5'-end of NHEIII_1 Element in human c-MYC promoter bound to triangulenium based fluorescence probe DAOTA-M2 | | Descriptor: | 8,12-bis(2-morpholinoethyl)-8H-benzo[ij]xantheno[1,9,8-cdef][2,7]naphthyridin-12-iumhexafluorophosphate, DNA (5'-D(*TP*AP*GP*GP*GP*AP*GP*GP*GP*TP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3') | | Authors: | Kotar, A, Wang, B, Shivalingam, A, Gonzalez-Garcia, J, Vilar, R, Plavec, J. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5LKN
 
 | | NMR solution structure of human FNIII domain 2 of NCAM | | Descriptor: | Neural cell adhesion molecule 1 | | Authors: | Slapsak, U, Salzano, G, Amin, L, Abskharon, R.N.N, Ilc, G, Zupancic, B, Biljan, I, Plavec, J, Giachin, G, Legname, G. | | Deposit date: | 2016-07-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The N Terminus of the Prion Protein Mediates Functional Interactions with the Neuronal Cell Adhesion Molecule (NCAM) Fibronectin Domain.
J.Biol.Chem., 291, 2016
|
|
5LM0
 
 | | NMR spatial structure of Tk-hefu peptide | | Descriptor: | L-2 | | Authors: | Mineev, K.S, Berkut, A.A, Novikova, E.V, Oparin, P.B, Grishin, E.V, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Protein Surface Topography as a tool to enhance the selective activity of a potassium channel blocker.
J.Biol.Chem., 2019
|
|
