4BZT
 
 | | The Solution Structure of the MLN 944-d(ATGCAT)2 Complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
4BZU
 
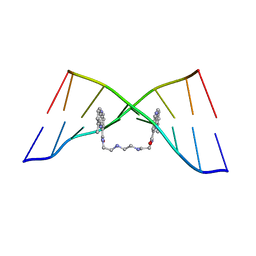 | | The Solution Structure of the MLN 944-d(TATGCATA)2 Complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
4BZV
 
 | | The Solution Structure of the MLN 944-d(TACGCGTA)2 complex | | Descriptor: | 1-METHYL-9-[12-(9-METHYLPHENAZIN-10-IUM-1-YL)-12-OXO-2,11-DIAZA-5,8-DIAZONIADODEC-1-ANOYL]PHENAZIN-10-IUM, DNA | | Authors: | Serobian, A, Thomas, D.S, Ball, G.E, Denny, W.A, Wakelin, L.P.G. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended.
Biopolymers, 101, 2014
|
|
4C26
 
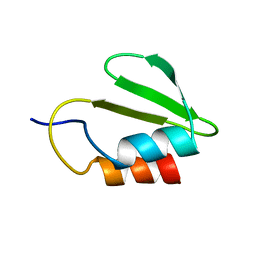 | | Solution NMR structure of the HicA toxin from Burkholderia pseudomallei | | Descriptor: | HICA | | Authors: | Butt, A, Higman, V.A, Williams, C, Crump, M.P, Hemsley, C, Harmer, N, Titball, R.W. | | Deposit date: | 2013-08-16 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Hica Toxin from Burkholderia Pseudomallei Has a Role in Persister Cell Formation.
Biochem.J., 459, 2014
|
|
4C7Q
 
 | |
4CA3
 
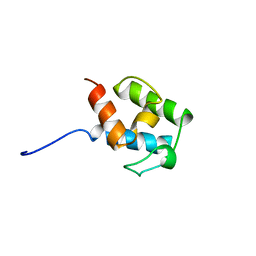 | | SOLUTION STRUCTURE OF STREPTOMYCES VIRGINIAE VIRA ACP5B | | Descriptor: | HYBRID POLYKETIDE SYNTHASE-NON RIBOSOMAL PEPTIDE SYNTHETASE | | Authors: | Davison, J, Dorival, J, Rabeharindranto, M.H, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2013-10-05 | | Release date: | 2014-06-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Insights Into the Function of Trans-Acyl Transferase Polyketide Synthases from the Saxs Structure of a Complete Module.
Chem.Sci., 2014
|
|
4CA9
 
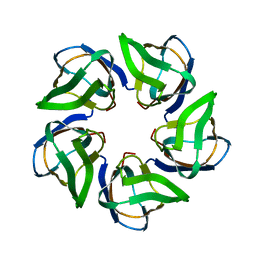 | | Structure of the Nucleoplasmin-like N-terminal domain of Drosophila FKBP39 | | Descriptor: | 39 KDA FK506-BINDING NUCLEAR PROTEIN | | Authors: | Artero, J, Forsyth, T, Callow, P, Watson, A.A, Zhang, W, Laue, E.D, Edlich-Muth, C, Przewloka, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Pentameric Nucleoplasmin Fold is Present in Drosophila Fkbp39 and a Large Number of Chromatin-Related Proteins.
J.Mol.Biol., 427, 2015
|
|
4CH0
 
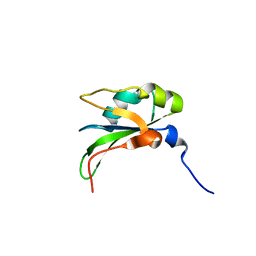 | | RRM domain from C. elegans SUP-12 | | Descriptor: | PROTEIN SUP-12, ISOFORM B | | Authors: | Amrane, S, Mackereth, C.D. | | Deposit date: | 2013-11-27 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Backbone-Independent Nucleic Acid Binding by Splicing Factor Sup-12 Reveals Key Aspects of Molecular Recognition
Nat.Commun., 5, 2014
|
|
4CH1
 
 | |
4CIO
 
 | |
4CPG
 
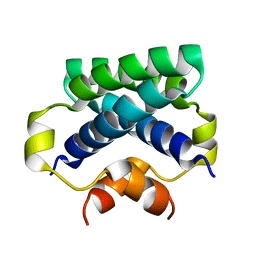 | |
4CRP
 
 | | Solution structure of a TrkAIg2 domain construct for use in drug discovery | | Descriptor: | HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR | | Authors: | Shoemark, D.K, Fahey, M, Williams, C, Sessions, R.B, Crump, M.P, Allen-Birt, S.J. | | Deposit date: | 2014-02-28 | | Release date: | 2015-01-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Design and Nuclear Magnetic Resonance (NMR) Structure Determination of the Second Extracellular Immunoglobulin Tyrosine Kinase a (Trkaig2) Domain Construct for Binding Site Elucidation in Drug Discovery
J.Med.Chem., 58, 2015
|
|
4CSQ
 
 | |
4CYK
 
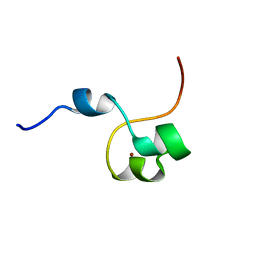 | | Structural basis for binding of Pan3 to Pan2 and its function in mRNA recruitment and deadenylation | | Descriptor: | PAB-DEPENDENT POLY(A)-SPECIFIC RIBONUCLEASE SUBUNIT PAN3, ZINC ION | | Authors: | Wolf, J, Valkov, E, Allen, M.D, Meineke, B, Gordiyenko, Y, McLaughlin, S.H, Olsen, T.M, Robinson, C.V, Bycroft, M, Stewart, M, Passmore, L.A. | | Deposit date: | 2014-04-11 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Pan3 Binding to Pan2 and its Function in Mrna Recruitment and Deadenylation.
Embo J., 33, 2014
|
|
4CZ3
 
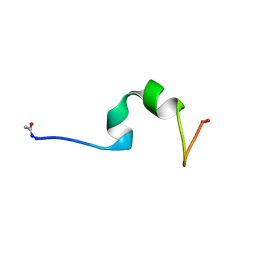 | | HP24wt derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
4CZ4
 
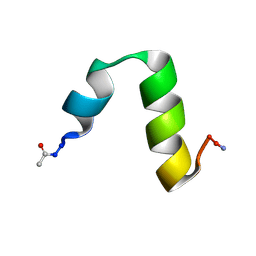 | | HP24stab derived from the villin headpiece subdomain | | Descriptor: | VILLIN-1 | | Authors: | Hocking, H, Haese, F, Madl, T, Zacharias, M, Rief, M, Zoldak, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-02-18 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Compact Native 24-Residue Supersecondary Structure Derived from the Villin Headpiece Subdomain.
Biophys.J., 108, 2015
|
|
4D4W
 
 | |
4D7X
 
 | |
4GAT
 
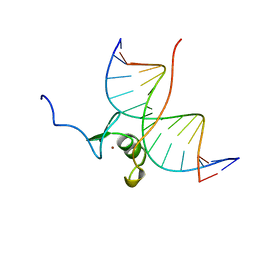 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
4UEI
 
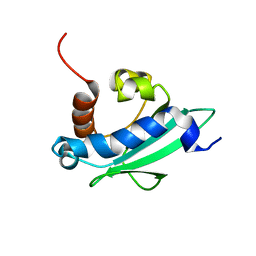 | | Solution structure of the sterol carrier protein domain 2 of Helicoverpa armigera | | Descriptor: | STEROL CARRIER PROTEIN 2/3-OXOACYL-COA THIOLASE | | Authors: | Liu, X, Ma, H, Yan, X, Hong, H, Peng, J, Peng, R. | | Deposit date: | 2014-12-18 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Function of Helicoverpa Armigera Sterol Carrier Protein-2, an Important Insecticidal Target from the Cotton Bollworm.
Sci.Rep., 5, 2015
|
|
4UET
 
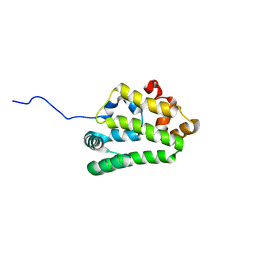 | | Diversity in the structures and ligand binding sites among the fatty acid and retinol binding proteins of nematodes revealed by Na-FAR-1 from Necator americanus | | Descriptor: | NEMATODE FATTY ACID RETINOID BINDING PROTEIN | | Authors: | Rey-Burusco, M.F, Ibanez Shimabukuro, M, Griffiths, K, Cooper, A, Kennedy, M.W, Corsico, B, Smith, B.O, Griffiths, K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Diversity in the Structures and Ligand Binding Sites of Nematode Fatty Acid and Retinol Binding Proteins Revealed by Na-Far-1 from Necator Americanus.
Biochem.J., 471, 2015
|
|
4ULL
 
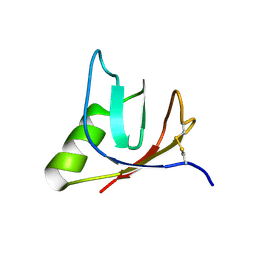 | | SOLUTION NMR STRUCTURE OF VEROTOXIN-1 B-SUBUNIT FROM E. COLI, 5 STRUCTURES | | Descriptor: | Shiga toxin 1B | | Authors: | Richardson, J.M, Evans, P.D, Homans, S.W, Donohue-Rolfe, A. | | Deposit date: | 1996-12-17 | | Release date: | 1997-04-01 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbohydrate-binding B-subunit homopentamer of verotoxin VT-1 from E. coli.
Nat.Struct.Biol., 4, 1997
|
|
4UQT
 
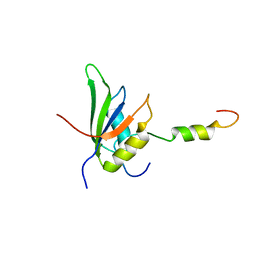 | | RRM-peptide structure in RES complex | | Descriptor: | PRE-MRNA-SPLICING FACTOR CWC26, U2 SNRNP COMPONENT IST3 | | Authors: | Tripsianes, K, Friberg, A, Barrandon, C, Seraphin, B, Sattler, M. | | Deposit date: | 2014-06-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel Protein-Protein Interaction in the Res (Retention and Splicing) Complex.
J.Biol.Chem., 289, 2014
|
|
4UZM
 
 | |
4UZW
 
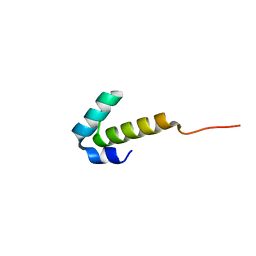 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
