5IEC
 
 | |
5IEJ
 
 | |
5IEM
 
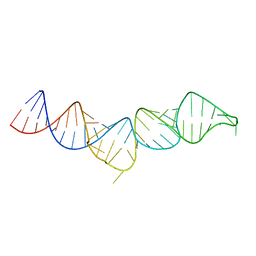 | | NMR structure of the 5'-terminal hairpin of the 7SK snRNA | | Descriptor: | 7SK snRNA | | Authors: | Bourbigot, S, Dock-Bregeon, A.C, Coutant, J, Kieffer, B, Lebars, I. | | Deposit date: | 2016-02-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA.
RNA, 22, 2016
|
|
5IEW
 
 | |
5IIR
 
 | |
5IIV
 
 | | GCN4p pH 1.5 | | Descriptor: | General control protein GCN4 | | Authors: | Brady, M.R, Kaplan, A.R, Alexandrescu, A.T. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structures of GCN4p Are Largely Conserved When Ion Pairs Are Disrupted at Acidic pH but Show a Relaxation of the Coiled Coil Superhelix.
Biochemistry, 56, 2017
|
|
5IJ4
 
 | | Solution structure of AN1-type zinc finger domain from Cuz1 (Cdc48 associated ubiquitin-like/zinc-finger protein-1) | | Descriptor: | CDC48-associated ubiquitin-like/zinc finger protein 1, ZINC ION | | Authors: | Sun, Z.-Y.J, Hanna, J, Wagner, G, Bhanu, M.K, Allan, M, Arthanari, H. | | Deposit date: | 2016-03-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cuz1 AN1 Zinc Finger Domain: An Exposed LDFLP Motif Defines a Subfamily of AN1 Proteins.
Plos One, 11, 2016
|
|
5IM8
 
 | |
5ION
 
 | |
5IPO
 
 | |
5IQ5
 
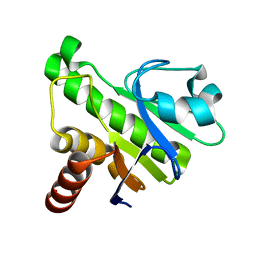 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
5IRD
 
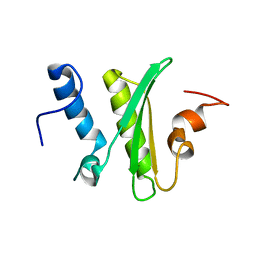 | |
5IRT
 
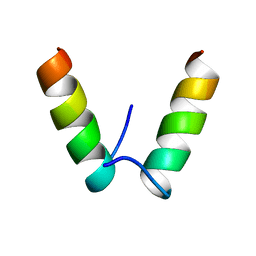 | |
5ISN
 
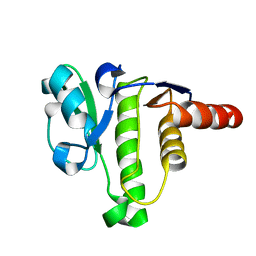 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus | | Descriptor: | Non-structural polyprotein | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Tsika, A.C, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-03-15 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
5IV1
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 4th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Gruber, D.R, Hoppins, J.J, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-18 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
5IX5
 
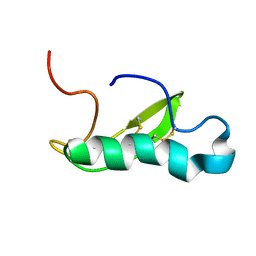 | | NMR structure of antibacterial factor-2 | | Descriptor: | Antibacterial factor-related peptide 2 | | Authors: | Masakatsu, K, Umetsu, Y, Rumi, F, Kikukawa, T, Ohki, S, Mizuguchi, M, Demura, M, Aizawa, T. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of ABF-2 (antibacterial factor-2) from C. elegans and the interaction with membrane mimetic systems
To Be Published
|
|
5IX9
 
 | | Cell surface anchoring domain | | Descriptor: | Antifreeze protein | | Authors: | Guo, S, Langelaan, D. | | Deposit date: | 2016-03-23 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice.
Sci Adv, 3, 2017
|
|
5IXF
 
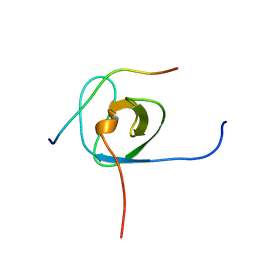 | | Solution structure of the STAM2 SH3 with AMSH derived peptide complex | | Descriptor: | STAM-binding protein, Signal transducing adapter molecule 2 | | Authors: | Hologne, M, Cantrelle, F.X, Trivelli, X, Walker, O. | | Deposit date: | 2016-03-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Reveals the Interplay among the AMSH SH3 Binding Motif, STAM2, and Lys63-Linked Diubiquitin.
J.Mol.Biol., 428, 2016
|
|
5IZB
 
 | |
5IZP
 
 | | Solution Structure of DNA Dodecamer with 8-oxoguanine at 10th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Kiryutin, A.S, Kasymov, R.D, Yurkovskaya, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-03-25 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 8-Oxoguanine Affects DNA Backbone Conformation in the EcoRI Recognition Site and Inhibits Its Cleavage by the Enzyme.
Plos One, 11, 2016
|
|
5J05
 
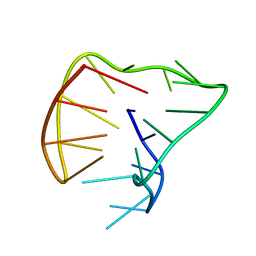 | |
5J0M
 
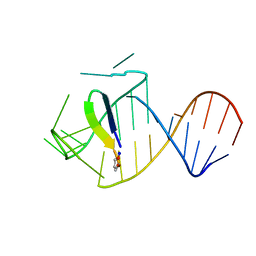 | | Ground state sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29-mer) of the HIV-1 TAR RNA element, Cyclic peptide mimetic of HIV-1 Tat | | Authors: | Borkar, A.N, Bardaro Jr, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-28 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J17
 
 | |
5J18
 
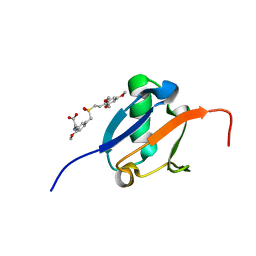 | | Solution structure of Ras Binding Domain (RBD) of B-Raf complexed with Rigosertib (Complex I) | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
5J1O
 
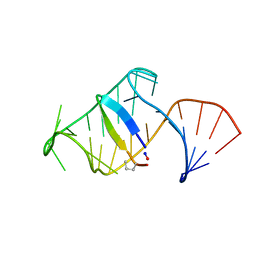 | | Excited state (Bound-like) sampled during RDC restrained Replica-averaged Metadynamics (RAM) simulations of the HIV-1 TAR complexed with cyclic peptide mimetic of Tat | | Descriptor: | Apical region (29mer) of the HIV-1 TAR element, Cyclic peptide mimetic of Tat | | Authors: | Borkar, A.N, Bardaro, M.F, Varani, G, Vendruscolo, M. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a low-population binding intermediate in protein-RNA recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
