4UZX
 
 | | High-resolution NMR structures of the domains of Saccharomyces cerevisiae Tho1 | | Descriptor: | PROTEIN THO1 | | Authors: | Jacobsen, J.O.B, Allen, M.D, Freund, S.M.V, Bycroft, M. | | Deposit date: | 2014-09-09 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution NMR Structures of the Domains of Saccharomyces Cerevisiae Tho1.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
4V10
 
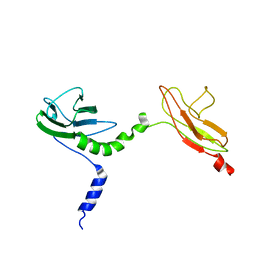 | | Skelemin Association with alpha2b,beta3 Integrin: A Structural Model | | Descriptor: | MYOMESIN-1 | | Authors: | Gorbatyuk, V, Deshmukh, L, Nguyen, K, Vinogradova, O. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Skelemin Association with Alphaiibbeta3 Integrin: A Structural Model.
Biochemistry, 53, 2014
|
|
5A17
 
 | |
5A18
 
 | |
5A3G
 
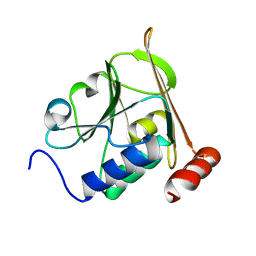 | | Structure of herpesvirus nuclear egress complex subunit M50 | | Descriptor: | M50 | | Authors: | Leigh, K.E, Boeszoermenyi, A, Mansueto, M.S, Sharma, M, Filman, D.J, Coen, D.M, Wagner, G, Hogle, J.M, Arthanari, H. | | Deposit date: | 2015-06-01 | | Release date: | 2015-07-15 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of a Herpesvirus Nuclear Egress Complex Subunit Reveals an Interaction Groove that is Essential for Viral Replication
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5A4G
 
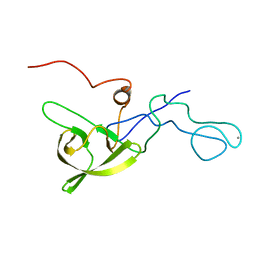 | | NMR structure of a 180 residue construct encompassing the N-terminal metal-binding site and the membrane proximal domain of SilB from Cupriavidus metallidurans CH34 | | Descriptor: | SILB, SILVER EFFLUX PROTEIN, MFP COMPONENT OF THE THREE COMPONENTS PROTON ANTIPORTER METAL EFFLUX SYSTEM, ... | | Authors: | Bersch, B, Urbina Fernandez, P, Vandenbussche, G. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Investigation of the Ag+/Cu+-Binding Domains of the Periplasmic Adaptor Protein Silb from Cupriavidus Metallidurans Ch34.
Biochemistry, 55, 2016
|
|
5A4H
 
 | | Solution structure of the lipid droplet anchoring peptide of CGI-58 bound to DPC micelles | | Descriptor: | 1-ACYLGLYCEROL-3-PHOSPHATE O-ACYLTRANSFERASE ABHD5 | | Authors: | Boeszoermenyi, A, Arthanari, H, Wagner, G, Nagy, H.M, Zangger, K, Lindermuth, H, Oberer, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Cgi-58 Motif Provides the Molecular Basis of Lipid Droplet Anchoring.
J.Biol.Chem., 290, 2015
|
|
5AAQ
 
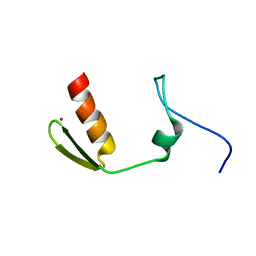 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | CALCIUM-BINDING AND COILED-COIL DOMAIN-CONTAINING PROTEIN 2, ZINC ION | | Authors: | Thurston, T.L, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-28 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
5AAS
 
 | | The selective autophagy receptor TAX1BP1 is required for autophagy- dependent capture of cytosolic Salmonella typhimurium | | Descriptor: | TAX1-BINDING PROTEIN 1, ZINC ION | | Authors: | Tumbarello, D.A, Manna, P.T, Allen, M, Bycroft, M, Kendrick-Jones, J, Buss, F. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Autophagy Receptor Tax1BP1 and the Molecular Motor Myosin Vi are Required for Clearance of Salmonella Typhimurium by Autophagy.
Plos Pathog., 11, 2015
|
|
5AAY
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | NF-KAPPA-B ESSENTIAL MODULATOR, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
5AAZ
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | OPTINEURIN, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
5ABK
 
 | | Structure of the N-terminal domain of the metalloprotease PrtV from Vibrio cholerae | | Descriptor: | METALLOPROTEASE | | Authors: | Persson, C, Mayzel, M, Edwin, A, Wai, S.N, Ohman, A, Sauer-Eriksson, A.E, Karlsson, G. | | Deposit date: | 2015-08-06 | | Release date: | 2015-08-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal Domain of the Metalloprotease Prtv from Vibrio Cholerae.
Protein Sci., 24, 2015
|
|
5AGQ
 
 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | Descriptor: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | Authors: | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | Deposit date: | 2015-02-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
5AHT
 
 | |
5AIW
 
 | |
5AJ1
 
 | | Solution Structure of the Smarc Domain | | Descriptor: | SWI/SNF-RELATED MATRIX-ASSOCIATED ACTIN-DEPENDENT REGULATOR OF CHROMATIN SUBFAMILY B MEMBER 1 | | Authors: | Allen, M.D, Freund, S.M.V, Zinzalla, G, Bycroft, M. | | Deposit date: | 2015-02-19 | | Release date: | 2015-08-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Swi/Snf Subunit Ini1 Contains an N-Terminal Winged Helix DNA Binding Domain that is a Target for Mutations in Schwannomatosis.
Structure, 23, 2015
|
|
5B7A
 
 | |
5B7J
 
 | | Structure model of Sap1-DNA complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*TP*TP*GP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*CP*AP*AP*TP*AP*TP*T)-3'), Switch-activating protein 1 | | Authors: | Jin, C, Hu, Y, Ding, J, Zhang, Y. | | Deposit date: | 2016-06-07 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sap1 is a replication-initiation factor essential for the assembly of pre-replicative complex in the fission yeast Schizosaccharomyces pombe
J. Biol. Chem., 292, 2017
|
|
5B7X
 
 | |
5B81
 
 | |
5B88
 
 | | RRM-like domain of DEAD-box protein, CsdA | | Descriptor: | ATP-dependent RNA helicase DeaD | | Authors: | Xu, L, Peng, J, Zhang, J, Wu, J, Tang, Y, Shi, Y. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the Structure of Dimeric RNA Helicase CsdA and Indispensable Role of Its C-Terminal Regions.
Structure, 25, 2017
|
|
5FRF
 
 | | Solution structure of reduced and zinc-bound RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA, ZINC ION | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
5FRG
 
 | |
5FRH
 
 | | Solution structure of oxidised RsrA | | Descriptor: | ANTI-SIGMA FACTOR RSRA | | Authors: | Zdanowski, K, Pecqueur, L, Werner, J, Potts, J.R, Kleanthous, C. | | Deposit date: | 2015-12-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Anti-Sigma Factor Rsra Responds to Oxidative Stress by Reburying its Hydrophobic Core.
Nat.Commun., 7, 2016
|
|
5FZV
 
 | |
