2W84
 
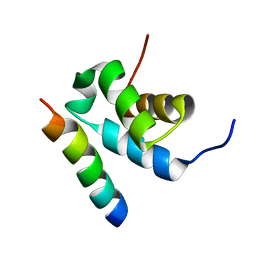 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
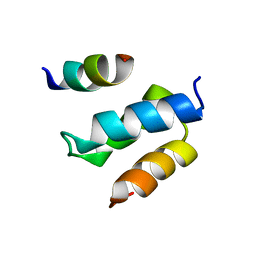 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
2W9O
 
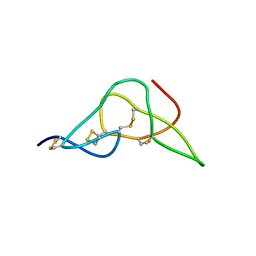 | | Solution structure of jerdostatin from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-27 | | Release date: | 2010-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9U
 
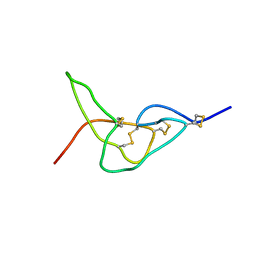 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9V
 
 | | Solution structure of jerdostatin from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2W9W
 
 | | Solution structure of jerdostatin mutant R24K from Trimeresurus jerdonii with end C-terminal residues N45G46 deleted | | Descriptor: | SHORT DISINTEGRIN JERDOSTATIN | | Authors: | Carbajo, R.J, Sanz, L, Mosulen, S, Calvete, J.J, Pineda-Lucena, A. | | Deposit date: | 2009-01-29 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Dynamics of Recombinant Wild-Type and Mutated Jerdostatin, a Selective Inhibitor of Integrin Alpha1 Beta1
Proteins, 79, 2011
|
|
2WBR
 
 | | The RRM domain in GW182 proteins contributes to miRNA-mediated gene silencing | | Descriptor: | GW182 | | Authors: | Eulalio, A, Tritschler, F, Buettner, R, Weichenrieder, O, Izaurralde, E, Truffault, V. | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Rrm Domain in Gw182 Proteins Contributes to Mirna-Mediated Gene Silencing.
Nucleic Acids Res., 37, 2009
|
|
2WC2
 
 | |
2WCC
 
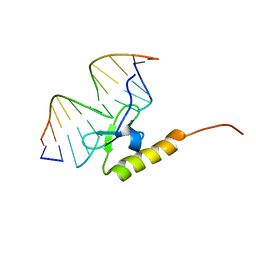 | | phage lambda IntDBD1-64 complex with p prime 2 DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DAP*DGP*DTP*DCP*DAP *DAP*DAP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DTP*DTP*DTP*DGP *DAP*DCP*DTP*DGP*DC)-3'), INTEGRASE | | Authors: | Fadeev, E.A, Sam, M.D, Clubb, R.T. | | Deposit date: | 2009-03-11 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Amino-Terminal Domain of the Lambda Integrase Protein in Complex with DNA: Immobilization of a Flexible Tail Facilitates Beta- Sheet Recognition of the Major Groove.
J.Mol.Biol., 388, 2009
|
|
2WCN
 
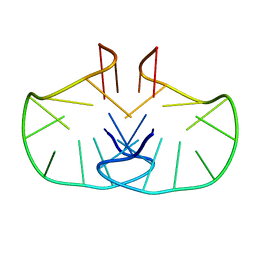 | | Solution structure of an LNA-modified quadruplex | | Descriptor: | DNA (5'-D(*DGP*LCG*DGP*LCG*DTP*DTP*DTP *DTP*DGP*LCG*DGP*LCG)-3') | | Authors: | Nielsen, J.T, Arar, K, Petersen, M. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Locked Nucleic Acid Modified Quadruplex: Introducing the V4 Folding Topology.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2WCY
 
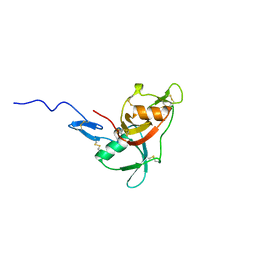 | | NMR solution structure of factor I-like modules of complement C7. | | Descriptor: | COMPLEMENT COMPONENT C7 | | Authors: | Phelan, M.M, Thai, C.T, Soares, D.C, Ogata, R.T, Barlow, P.N, Bramham, J. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Factor I-Like Modules from Complement C7 Reveals a Pair of Follistatin Domains in Compact Pseudosymmetric Arrangement.
J.Biol.Chem., 284, 2009
|
|
2WGN
 
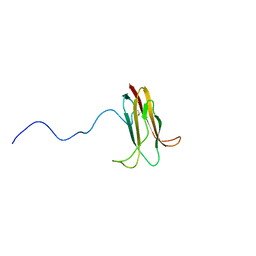 | | Pseudomonas aeruginosa ICP | | Descriptor: | INHIBITOR OF CYSTEINE PEPTIDASE COMPND 3 | | Authors: | Fu, Y, Smith, B.O. | | Deposit date: | 2009-04-21 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Pseudomonas Aeruginosa Icp
Ph D Thesis, 2009
|
|
2WGO
 
 | | Structure of ranaspumin-2, a surfactant protein from the foam nests of a tropical frog | | Descriptor: | RANASPUMIN-2 | | Authors: | Mackenzie, C.D, Smith, B.O, Kennedy, M.W, Cooper, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-06-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Ranaspumin-2: Structure and Function of a Surfactant Protein from the Foam Nests of a Tropical Frog.
Biophys.J., 96, 2009
|
|
2WH9
 
 | | Solution structure of GxTX-1E | | Descriptor: | GUANGXITOXIN-1EGXTX-1E | | Authors: | Lee, S.K, Jung, H.H, Lee, J.Y, Lee, C.W, Kim, J.I. | | Deposit date: | 2009-05-02 | | Release date: | 2010-05-26 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Gxtx-1E, a High Affinity Tarantula Toxin Interacting with Voltage Sensors in Kv2.1 Potassium Channels.
Biochemistry, 49, 2010
|
|
2WNM
 
 | | Solution structure of Gp2 | | Descriptor: | GENE 2 | | Authors: | Camara, B, Liu, M, Shadrinc, A, Liu, B, Simpson, P, Weinzierl, R, Severinovc, K, Cota, E, Matthews, S, Wigneshweraraj, S.R. | | Deposit date: | 2009-07-13 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | T7 Phage Protein Gp2 Inhibits the Escherichia Coli RNA Polymerase by Antagonizing Stable DNA Strand Separation Near the Transcription Start Site.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WQG
 
 | | SAP domain from Tho1: L31W (fluorophore) mutant | | Descriptor: | PROTEIN THO1 | | Authors: | Dodson, C.A, Ferguson, N, Rutherford, T.J, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2009-08-21 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Engineering a Two-Helix Bundle Protein for Folding Studies.
Protein Eng.Des.Sel., 23, 2010
|
|
2WWV
 
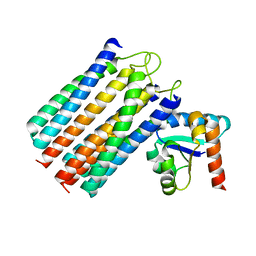 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N\,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N\,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2WXC
 
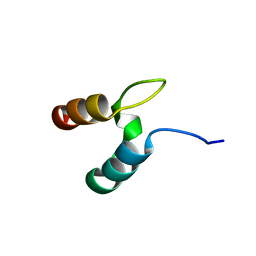 | | The folding mechanism of BBL: Plasticity of transition-state structure observed within an ultrafast folding protein family. | | Descriptor: | DIHYDROLIPOYLTRANSSUCCINASE | | Authors: | Neuweiler, H, Sharpe, T.D, Rutherford, T.J, Johnson, C.M, Allen, M.D, Ferguson, N, Fersht, A.R. | | Deposit date: | 2009-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Folding Mechanism of Bbl: Plasticity of Transition-State Structure Observed within an Ultrafast Folding Protein Family.
J.Mol.Biol., 390, 2009
|
|
2WY2
 
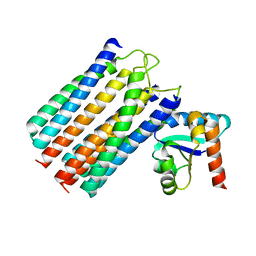 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N\,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N\,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
2X43
 
 | | STRUCTURAL BASIS OF MOLECULAR RECOGNITION BY SHERP AT MEMBRANE SURFACES | | Descriptor: | SHERP | | Authors: | Moore, B, Miles, A.J, Guerra, C.G, Simpson, P, Iwata, M, Wallace, B.A, Matthews, S.J, Smith, D.F, Brown, K.A. | | Deposit date: | 2010-02-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Moelcular Recognition by the Leishmania Small Hydrophilic Endoplasmic Reticulum-Associated Protein, Sherp, at Membrane Surfaces
J.Biol.Chem., 286, 2011
|
|
2X8N
 
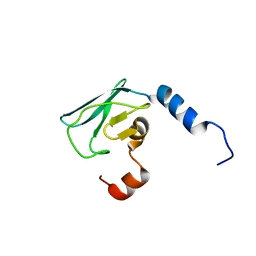 | | Solution NMR structure of uncharacterized protein CV0863 from Chromobacterium violaceum. NORTHEAST STRUCTURAL GENOMICS TARGET (NESG) target CvT3. OCSP target CV0863. | | Descriptor: | CV0863 | | Authors: | Gutmanas, A, Fares, C, Yee, A, Lemak, A, Semesi, A, Arrowsmith, C.H, Ontario Centre for Structural Proteomics (OCSP), Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Uncharacterized Protein Cv0863 from Chromobacterium Violaceum
To be Published
|
|
2XA6
 
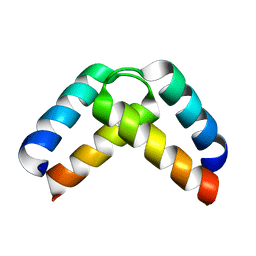 | | Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain | | Descriptor: | KH DOMAIN-CONTAINING\,RNA-BINDING\,SIGNAL TRANSDUCTION-ASSOCIATED PROTEIN 1 | | Authors: | Meyer, N.H, Tripsianes, K, Vincendeaux, M, Madl, T, Kateb, F, Brack-Werner, R, Sattler, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain.
J. Biol. Chem., 285, 2010
|
|
2XBD
 
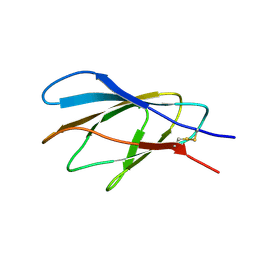 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-27 | | Release date: | 1999-07-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
2XC7
 
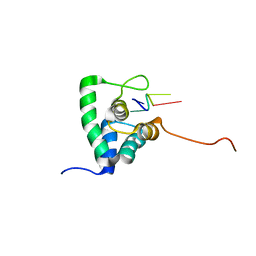 | | Solution Structure of PHAX-RBD in complex with ssRNA | | Descriptor: | 5'-(AP*UP*CP*GP)-3', PHOSPHORYLATED ADAPTER RNA EXPORT PROTEIN | | Authors: | Mourao, A, Mackereth, C.D, Varrot, A, Cusack, S, Sattler, M. | | Deposit date: | 2010-04-18 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and RNA Recognition by the Snrna and Snorna Transport Factor Phax.
RNA, 16, 2010
|
|
2XDF
 
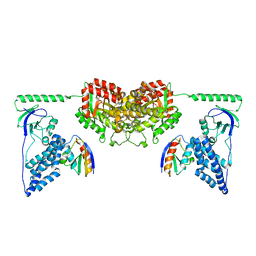 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | Authors: | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | Deposit date: | 2010-04-30 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
