2KXE
 
 | |
2KXF
 
 | | Solution structure of the first two RRM domains of FBP-interacting repressor (FIR) | | Descriptor: | Poly(U)-binding-splicing factor PUF60 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KXG
 
 | | The solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) | | Descriptor: | Aspartic protease inhibitor | | Authors: | Headey, S.J, Macaskill, U.K, Wright, M, Claridge, J.K, Edwards, P.J.B, Farley, P.C, Christeller, J.T, Laing, W.A, Pascal, S.M. | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the squash aspartic acid proteinase inhibitor (SQAPI) and mutational analysis of pepsin inhibition.
J.Biol.Chem., 285, 2010
|
|
2KXH
 
 | | Solution structure of the first two RRM domains of FIR in the complex with FBP Nbox peptide | | Descriptor: | Poly(U)-binding-splicing factor PUF60, peptide of Far upstream element-binding protein 1 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KXI
 
 | | Solution NMR structure of the apoform of NarE (NMB1343) | | Descriptor: | Uncharacterized protein | | Authors: | Koehler, C, Carlier, L, Veggi, D, Soriani, M, Pizza, M, Boelens, R, Bonvin, A.M.J.J. | | Deposit date: | 2010-05-06 | | Release date: | 2011-03-02 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the apoform of NarE (NMB1343)
To be Published
|
|
2KXJ
 
 | | Solution structure of UBX domain of human UBXD2 protein | | Descriptor: | UBX domain-containing protein 4 | | Authors: | Wu, Q, Huang, H, Zhang, J, Hu, Q, Wu, J, Shi, Y. | | Deposit date: | 2010-05-06 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution strcture of UBX domain of human UBXD2 protein
To be Published
|
|
2KXK
 
 | | Human Insulin Mutant A22Gly-B31Lys-B32Arg | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Bocian, W, Kozerski, L. | | Deposit date: | 2010-05-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Novel recombinant insulin analogue with flexible C - terminus in B chain. NMR structure of biosynthetic engineered A22G-B31K-B32R human insulin monomer in water /acetonitrile solution.
To be Published
|
|
2KXL
 
 | |
2KXM
 
 | | Solution NMR Structure of the 27 nucleotide engineered neomycin sensing riboswitch RNA-ribostmycin complex | | Descriptor: | RIBOSTAMYCIN, RNA (27-MER) | | Authors: | Duchardt-Ferner, E, Weigand, J.E, Ohlenschlager, O, Schmidtke, S.R, Suess, B, Wohnert, J. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Highly modular structure and ligand binding by conformational capture in a minimalistic riboswitch.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2KXN
 
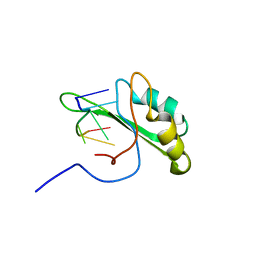 | | NMR structure of human Tra2beta1 RRM in complex with AAGAAC RNA | | Descriptor: | 5'-R(*AP*AP*GP*AP*AP*C)-3', Transformer-2 protein homolog beta | | Authors: | Clery, A, Jayne, S, Benderska, N, Dominguez, C, Stamm, S, Allain, F.H.-T. | | Deposit date: | 2010-05-10 | | Release date: | 2011-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1
Nat.Struct.Mol.Biol., 18, 2011
|
|
2KXO
 
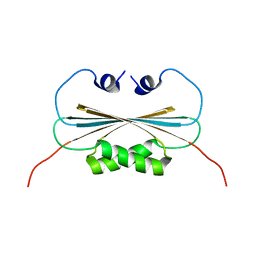 | |
2KXP
 
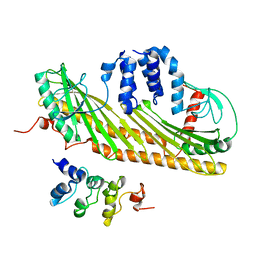 | |
2KXQ
 
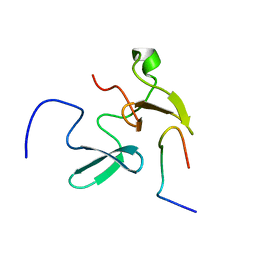 | | Solution Structure of Smurf2 WW2 and WW3 bound to Smad7 PY motif containing peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF2, Smad7 PY motif containing peptide | | Authors: | Chong, A, Lin, H, Wrana, J, Forman-Kay, J.D. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2KXR
 
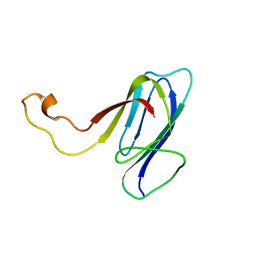 | | ZO1 ZU5 domain MC/AA mutation | | Descriptor: | Tight junction protein ZO-1 | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cdc42-dependent formation of the ZO-1/MRCKb complex at the leading edge controls cell migration
Embo J., 30, 2011
|
|
2KXS
 
 | | ZO1 ZU5 domain in complex with GRINL1A peptide | | Descriptor: | Tight junction protein ZO-1,Myocardial zonula adherens protein | | Authors: | Wen, W, Zhang, M. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-30 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Cdc42-dependent formation of the ZO-1/MRCKb complex at the leading edge controls cell migration
Embo J., 30, 2011
|
|
2KXT
 
 | |
2KXV
 
 | |
2KXW
 
 | |
2KXX
 
 | | NMR Structure of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex | | Descriptor: | Small protein A | | Authors: | Kim, K, Okon, M, Escobar, E, Kang, H, McIntosh, L, Paetzel, M. | | Deposit date: | 2010-05-13 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Escherichia coli BamE, a Lipoprotein Component of the beta-Barrel Assembly Machinery Complex.
Biochemistry, 50, 2011
|
|
2KXY
 
 | | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C | | Descriptor: | Uncharacterized protein | | Authors: | Liu, Y, Lee, H, Belote, R, Ciccosanti, C, Hamilton, K, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SuR18C from Streptococcus thermophilus. Northeast Structural Genomics Consortium Target SuR18C
To be Published
|
|
2KXZ
 
 | |
2KY0
 
 | |
2KY1
 
 | |
2KY2
 
 | |
2KY3
 
 | |
