1RQV
 
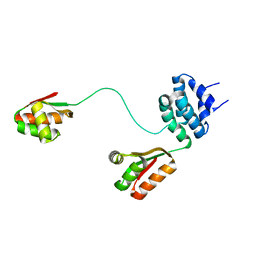 | | Spatial model of L7 dimer from E.coli with one hinge region in helical state | | Descriptor: | 50S ribosomal protein L7/L12 | | Authors: | Bocharov, E.V, Sobol, A.G, Pavlov, K.V, Korzhnev, D.M, Jaravine, V.A, Gudkov, A.T, Arseniev, A.S. | | Deposit date: | 2003-12-07 | | Release date: | 2004-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | From structure and dynamics of protein L7/L12 to molecular switching in ribosome
J.Biol.Chem., 279, 2004
|
|
1RRD
 
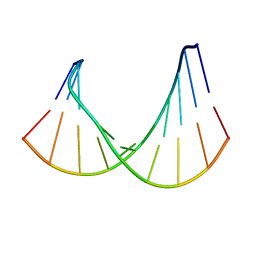 | | DNA/RNA HYBRID DUPLEX CONTAINING A PURINE-RICH RNA STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*CP*TP*CP*TP*TP*C)-3'), RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1RRR
 
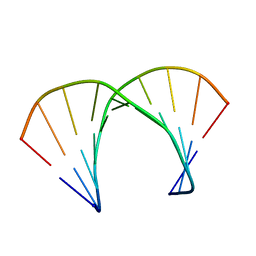 | | RNA DUPLEX CONTAINING A PURINE-RICH STRAND, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*AP*AP*GP*AP*GP*AP*AP*GP*C)-3'), RNA (5'-R(*GP*CP*UP*UP*CP*UP*CP*UP*UP*C)-3') | | Authors: | Gyi, J.I, Lane, A.N, Conn, G.L, Brown, T. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of DNA.RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes.
Biochemistry, 37, 1998
|
|
1RSF
 
 | |
1RSO
 
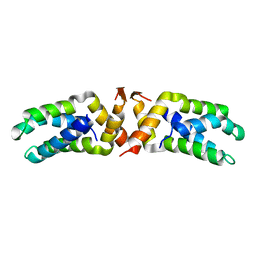 | | Hetero-tetrameric L27 (Lin-2, Lin-7) domain complexes as organization platforms of supra-molecular assemblies | | Descriptor: | Peripheral plasma membrane protein CASK, Presynaptic protein SAP97 | | Authors: | Feng, W, Long, J.-F, Fan, J.-S, Suetake, T, Zhang, M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The tetrameric L27 domain complex as an organization platform for supramolecular assemblies
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
1RSW
 
 | | 12-mer from site II calbindin D9K (DKNGDGEVSFEE) coordination Pb(II) | | Descriptor: | Vitamin D-dependent calcium-binding protein, intestinal | | Authors: | Ferrari, R, Mendoza, G, James, T, Tonelli, M, Basus, V, Gasque, L. | | Deposit date: | 2003-12-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Coordination Compounds derived from a dodecapeptide and Pb2+, Cd2+ and Zn2+. Modeling the site II from the Calbindin D9K
To be published
|
|
1RTN
 
 | |
1RU5
 
 | |
1RUU
 
 | |
1RVH
 
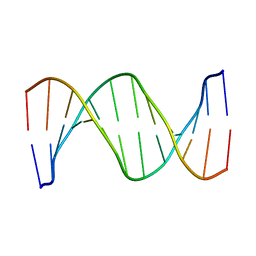 | | SOLUTION STRUCTURE OF THE DNA DODECAMER GCAAAATTTTGC | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*AP*TP*TP*TP*TP*GP*C)-3' | | Authors: | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | Deposit date: | 2003-12-13 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RVI
 
 | | SOLUTION STRUCTURE OF THE DNA DODECAMER CGTTTTAAAACG | | Descriptor: | 5'-D(*CP*GP*TP*TP*TP*TP*AP*AP*AP*AP*CP*G)-3' | | Authors: | Stefl, R, Wu, H, Ravindranathan, S, Sklenar, V, Feigon, J. | | Deposit date: | 2003-12-13 | | Release date: | 2004-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | DNA A-tract bending in three dimensions: Solving the dA4T4 vs. dT4A4 conundrum.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RVS
 
 | | STRUCTURE OF TRANSTHYRETIN IN AMYLOID FIBRILS DETERMINED BY SOLID-STATE MAGIC ANGLE SPINNING NMR | | Descriptor: | Transthyretin | | Authors: | Jaroniec, C.P, Macphee, C.E, Bajaj, V.S, Mcmahon, M.T, Dobson, C.M, Griffin, R.G. | | Deposit date: | 2003-12-14 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
1RW5
 
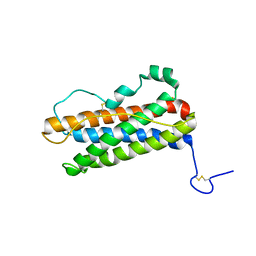 | |
1RWS
 
 | |
1RWU
 
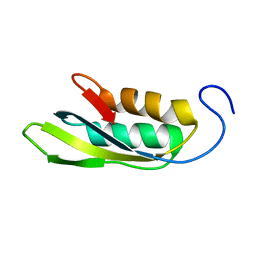 | |
1RXL
 
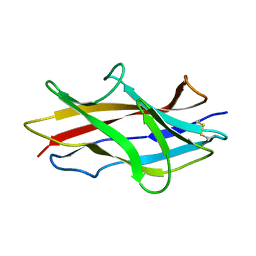 | | Solution structure of the engineered protein Afae-dsc | | Descriptor: | Afimbrial adhesin AFA-III | | Authors: | Anderson, K.L, Billington, J, Pettigrew, D, Cota, E, Roversi, P, Simpson, P, Chen, H.A, Urvil, P, du Merle, L, Barlow, P.N, Medof, M.E, Smith, R.A, Nowicki, B, Le Bouguenec, C, Lea, S.M, Matthews, S. | | Deposit date: | 2003-12-18 | | Release date: | 2005-01-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | An atomic resolution model for assembly, architecture, and function of the Dr adhesins.
Mol.Cell, 15, 2004
|
|
1RXR
 
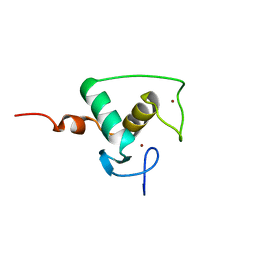 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE RETINOID X RECEPTOR DNA BINDING DOMAIN, NMR, 20 STRUCTURE | | Descriptor: | RETINOIC ACID RECEPTOR-ALPHA, ZINC ION | | Authors: | Holmbeck, S.M.A, Foster, M.P, Casimiro, D.R, Sem, D.S, Dyson, H.J, Wright, P.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the retinoid X receptor DNA-binding domain.
J.Mol.Biol., 281, 1998
|
|
1RY3
 
 | | NMR Solution Structure of the Precursor for Carnobacteriocin B2, an Antimicrobial Peptide from Carnobacterium piscicola | | Descriptor: | Bacteriocin carnobacteriocin B2 | | Authors: | Sprules, T, Kawulka, K.E, Gibbs, A.C, Wishart, D.S, Vederas, J.C. | | Deposit date: | 2003-12-19 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the precursor for carnobacteriocin B2, an antimicrobial peptide from Carnobacterium piscicola.
Eur.J.Biochem., 271, 2004
|
|
1RY4
 
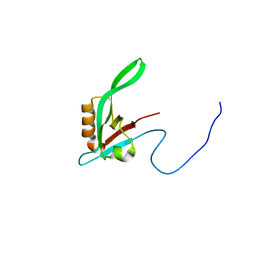 | |
1RYG
 
 | |
1RYJ
 
 | | Solution NMR Structure of Protein Mth1743 from Methanobacterium thermoautotrophicum. Ontario Centre for Structural Proteomics target MTH1743_1_70; Northeast Structural Genomics Consortium Target TT526. | | Descriptor: | unknown | | Authors: | Yee, A, Chang, X, Pineda-Lucena, A, Wu, B, Semesi, A, Le, B, Ramelot, T, Lee, G.M, Bhattacharyya, S, Gutierrez, P, Denisov, A, Lee, C.H, Cort, J.R, Kozlov, G, Liao, J, Finak, G, Chen, L, Wishart, D, Lee, W, McIntosh, L.P, Gehring, K, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | AN NMR APPROACH TO STRUCTURAL PROTEOMICS
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1RYK
 
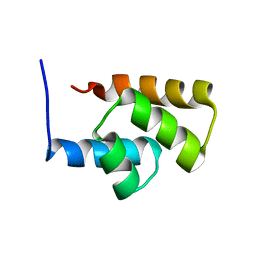 | | Solution NMR Structure Protein yjbJ from Escherichia coli. Northeast Structural Genomics Consortium Target ET93; Ontario Centre for Structural Proteomics target EC0298_1_69; | | Descriptor: | Protein yjbJ | | Authors: | Pineda-Lucena, A, Liao, J, Wu, B, Yee, A, Cort, J.R, Kennedy, M.A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-12-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proteins, 47, 2002
|
|
1RYV
 
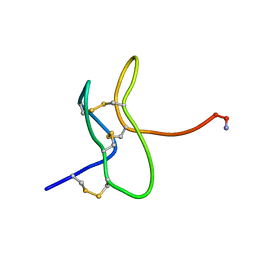 | |
1RZS
 
 | |
