8DPX
 
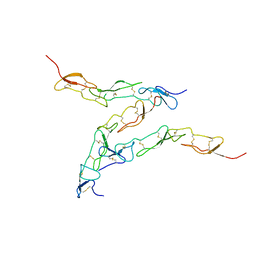 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
8DRH
 
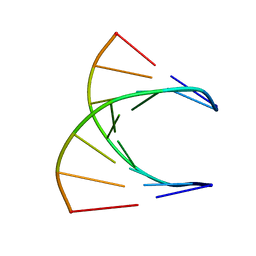 | | HIGH RESOLUTION NMR STRUCTURE OF THE D(GCGTCAGG)R(CCUGACGC) HYBRID, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*GP*TP*CP*AP*GP*G)-3'), RNA (5'-R(*CP*CP*UP*GP*AP*CP*GP*C)-3') | | Authors: | Bachelin, M, Hessler, G, Kurz, G, Hacia, J.G, Dervan, P.B, Kessler, H. | | Deposit date: | 1997-10-13 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stereoregular Phosphorothioate DNA/RNA Duplex
Nat.Struct.Biol., 5, 1998
|
|
8DSB
 
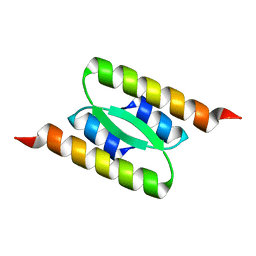 | | Lambda Bacteriophage Orf63 | | Descriptor: | Xis (Excision72) | | Authors: | Donaldson, L.W. | | Deposit date: | 2022-07-22 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Bad Phages in Good Bacteria: Role of the Mysterious orf63 of lambda and Shiga Toxin-Converting Phi 24 B Bacteriophages.
Front Microbiol, 8, 2017
|
|
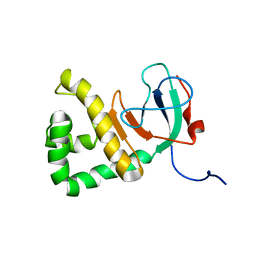
8DSX
 
 | |
8DWQ
 
 | |
8DYM
 
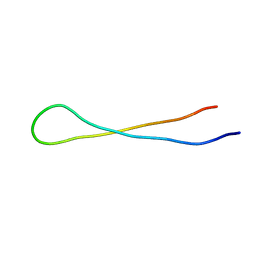 | | Aspartimidylated Graspetide Amycolimiditide | | Descriptor: | ATP-grasp target RiPP | | Authors: | Link, A.J, Choi, B. | | Deposit date: | 2022-08-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Mechanistic Analysis of the Biosynthesis of the Aspartimidylated Graspetide Amycolimiditide.
J.Am.Chem.Soc., 144, 2022
|
|
8DYN
 
 | |
8E1D
 
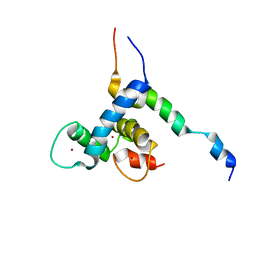 | |
8E22
 
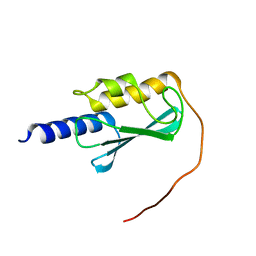 | | VPS37A_21-148 | | Descriptor: | Vacuolar protein sorting-associated protein 37A | | Authors: | Tian, F, Ye, Y.S. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Identification of membrane curvature sensing motifs essential for VPS37A phagophore recruitment and autophagosome closure.
Commun Biol, 7, 2024
|
|
8E2O
 
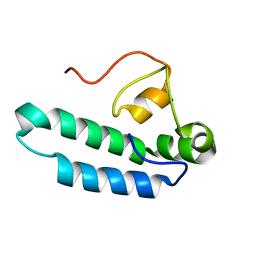 | |
8E4V
 
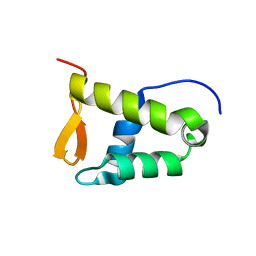 | | Solution structure of the WH domain of MORF | | Descriptor: | Isoform 3 of Histone acetyltransferase KAT6B | | Authors: | Zhang, Y, Kutateladze, T.G. | | Deposit date: | 2022-08-19 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | MORF and MOZ acetyltransferases target unmethylated CpG islands through the winged helix domain.
Nat Commun, 14, 2023
|
|
8E6Y
 
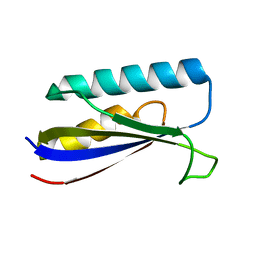 | |
8EB1
 
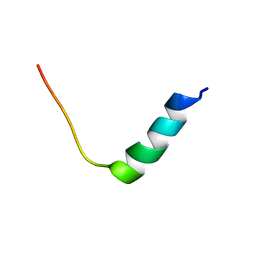 | | Chim2 - Intragenic antimicrobial peptide | | Descriptor: | Unconventional myosin-Ih, Transcription activator BRG1 intragenic antimicrobial chimeric peptide | | Authors: | de Freitas, T.V, Oliveira, A.L, Santos, M.A, Brand, G.D. | | Deposit date: | 2022-08-30 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Release of immunomodulatory peptides at bacterial membrane interfaces as a novel strategy to fight microorganisms.
J.Biol.Chem., 299, 2023
|
|
8EF4
 
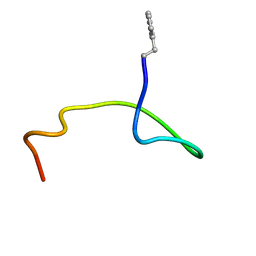 | |
8ENP
 
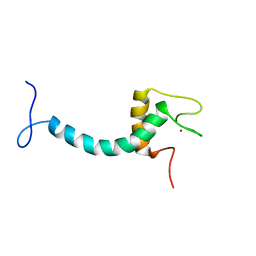 | | UBE3A isoform 3 AZUL | | Descriptor: | Isoform III of Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Bregnard, T.A, Bezsonova, I. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differences in structure, dynamics and Zn-coordination between isoforms of human ubiquitin ligase UBE3A
To Be Published
|
|
8EO9
 
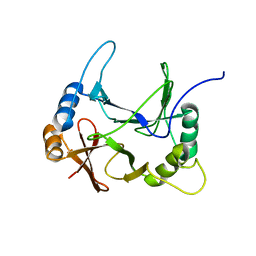 | | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX | | Descriptor: | Glyoxalase | | Authors: | Jia, X, Yan, X, Mobli, M, Qu, X. | | Deposit date: | 2022-10-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
To Be Published
|
|
8EOD
 
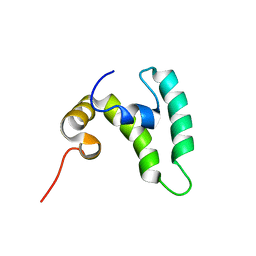 | |
8EP5
 
 | | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R1 | | Descriptor: | Mastoparan-R1 peptide | | Authors: | Freitas, C.D.P, Oshiro, K.G.N, Macedo, M.L.R, Cardoso, M.H, Franco, O.L, Liao, L.M. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-18 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R1.
To Be Published
|
|
8EPT
 
 | | UBE3A isoform 2 AZUL domain | | Descriptor: | Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Bregnard, T.A, Bezsonova, I. | | Deposit date: | 2022-10-06 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Differences in structure, dynamics and Zn-coordination between isoforms of human ubiquitin ligase UBE3A
To Be Published
|
|
8EPY
 
 | | The solution structure of abxF in complex with its product (-)-ABX, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | (6R,16R)-3,11,13,15-tetrahydroxy-1,6,9,9-tetramethyl-6,7,9,16-tetrahydro-14H-6,16-epoxyanthra[2,3-e]benzo[b]oxocin-14-one, Glyoxalase | | Authors: | Jia, X, Yan, X, Qu, X, Mobli, M. | | Deposit date: | 2022-10-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of abxF, an enzyme catalyzing the formation of chiral spiroketal of an antibiotics, (-)-ABX.
To Be Published
|
|
8ERU
 
 | | Solution NMR structure of a computationally designed mastoparan-like peptide, mastoparan-R4 | | Descriptor: | Mastoparan-R4 peptide | | Authors: | Freitas, C.D.P, Oshiro, K.G.N, Macedo, M.L.R, Cardoso, M.H, Franco, O.L, Liao, L.M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-18 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a computacionally designed mastoparan-like peptide, mastoparan-R4
To Be Published
|
|
8ERY
 
 | |
8ERZ
 
 | |
8ES0
 
 | |
8ES1
 
 | |
