1K1V
 
 | | Solution Structure of the DNA-Binding Domain of MafG | | Descriptor: | MafG | | Authors: | Kusunoki, H, Motohashi, H, Katsuoka, F, Morohashi, A, Yamamoto, M, Tanaka, T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA-binding domain of MafG.
Nat.Struct.Biol., 9, 2002
|
|
1K1Z
 
 | | Solution structure of N-terminal SH3 domain mutant(P33G) of murine Vav | | Descriptor: | vav | | Authors: | Ogura, K, Nagata, K, Horiuchi, M, Ebisui, E, Hasuda, T, Yuzawa, S, Nishida, M, Hatanaka, H, Inagaki, F. | | Deposit date: | 2001-09-26 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of N-terminal SH3 domain of Vav and the recognition site for Grb2 C-terminal SH3 domain
J.BIOMOL.NMR, 22, 2002
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
1K2H
 
 | | Three-dimensional Solution Structure of apo-S100A1. | | Descriptor: | S-100 protein, alpha chain | | Authors: | Rustandi, R.R, Baldisseri, D.M, Inman, K.G, Nizner, P, Hamilton, S.M, Landar, A, Landar, A, Zimmer, D.B, Weber, D.J. | | Deposit date: | 2001-09-27 | | Release date: | 2002-02-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the calcium-signaling protein apo-S100A1 as determined by NMR.
Biochemistry, 41, 2002
|
|
1K2J
 
 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-27 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
1K2K
 
 | | NMR MINIMIZED AVERAGE STRUCTURE OF d(CGTACG)2 | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Lam, S.L, Ip, L.N. | | Deposit date: | 2001-09-28 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Low temperature solution structures and base pair stacking of double helical d(CGTACG)(2).
J.Biomol.Struct.Dyn., 19, 2002
|
|
1K2M
 
 | |
1K2N
 
 | |
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3J
 
 | | Refined NMR Structure of the FHA1 Domain of Yeast Rad53 | | Descriptor: | Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3M
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALA, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | INSULIN | | Authors: | Xu, B, Hua, Q.-X, Nakagawa, S.H, Jia, W, Chu, Y.-C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A cavity-forming mutation in insulin induces segmental unfolding of a surrounding alpha-helix.
Protein Sci., 11, 2002
|
|
1K3N
 
 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3Q
 
 | | NMR structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T192) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K42
 
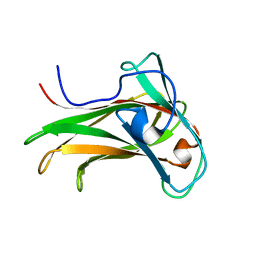 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
1K43
 
 | | 10 Structure Ensemble of the 14-residue peptide RG-KWTY-NG-ITYE-GR (MBH12) | | Descriptor: | MBH12 | | Authors: | Pastor, M.T, Lopez de la Paz, M, Lacroix, E, Serrano, L, Perez-Paya, E. | | Deposit date: | 2001-10-05 | | Release date: | 2001-10-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K45
 
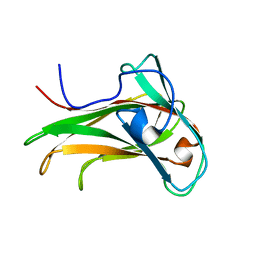 | | The Solution Structure of the CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase. | | Descriptor: | Xylanase | | Authors: | Simpson, P.J, Jamieson, S.J, Abou-Hachem, M, Nordberg-Karlsson, E, Gilbert, H.J, Holst, O, Williamson, M.P. | | Deposit date: | 2001-10-05 | | Release date: | 2002-05-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the CBM4-2 carbohydrate binding module from a thermostable Rhodothermus marinus xylanase.
Biochemistry, 41, 2002
|
|
1K4U
 
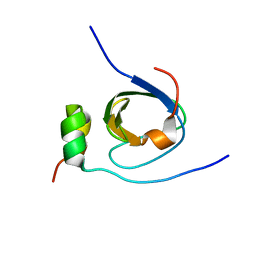 | | Solution structure of the C-terminal SH3 domain of p67phox complexed with the C-terminal tail region of p47phox | | Descriptor: | PHAGOCYTE NADPH OXIDASE SUBUNIT P47PHOX, PHAGOCYTE NADPH OXIDASE SUBUNIT P67PHOX | | Authors: | Kami, K, Takeya, R, Sumimoto, H, Kohda, D. | | Deposit date: | 2001-10-08 | | Release date: | 2002-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Diverse recognition of non-PxxP peptide ligands by the SH3 domains from p67(phox), Grb2 and Pex13p.
EMBO J., 21, 2002
|
|
1K5O
 
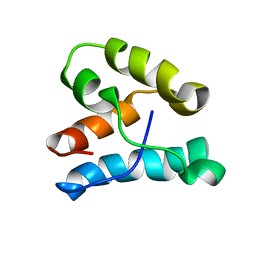 | | CPI-17(35-120) deletion mutant | | Descriptor: | CPI-17 | | Authors: | Ohki, S, Eto, M, Kariya, E, Hayano, T, Hayashi, Y, Yazawa, M, Brautigan, D, Kainosho, M. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Myosin Phosphatase Inhibitor Protein CPI-17 Shows Phosphorylation-induced Conformational Changes Responsible for Activation
J.Mol.Biol., 314, 2001
|
|
1K5R
 
 | | YAP65 WW domain S24-Amino-Ethylsulfanyl-Acetic Acid mutant | | Descriptor: | 65 KDA YES-ASSOCIATED PROTEIN, Fragment of WBP-1 | | Authors: | Ferguson, N, Pires, J.R, Toepert, F, Johnson, C.M, Pan, Y.P, Volkmer-Engert, R, Schneider-Mergener, J, Daggett, V, Oschkinat, H, Fersht, A.R. | | Deposit date: | 2001-10-12 | | Release date: | 2001-11-02 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Using flexible loop mimetics to extend phi-value analysis to secondary structure interactions.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1K5W
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE SYNAPTOTAGMIN 1 C2B-DOMAIN: SYNAPTOTAGMIN 1 AS A PHOSPHOLIPID BINDING MACHINE | | Descriptor: | CALCIUM ION, Synaptotagmin I | | Authors: | Fernandez, I, Arac, D, Ubach, J, Gerber, S.H, Shin, O, Gao, Y, Anderson, R.G.W, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-10-12 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the synaptotagmin 1 C2B-domain: synaptotagmin 1 as a phospholipid binding machine.
Neuron, 32, 2001
|
|
1K64
 
 | | NMR Structue of alpha-conotoxin EI | | Descriptor: | alpha-conotoxin EI | | Authors: | Park, K.H, Suk, J.E, Jacobsen, R, Gray, W.R, McIntosh, J.M, Han, K.H. | | Deposit date: | 2001-10-15 | | Release date: | 2003-09-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of alpha-conotoxin EI, a neuromuscular toxin specific for the alpha 1/delta subunit interface of torpedo nicotinic acetylcholine receptor
J.BIOL.CHEM., 276, 2001
|
|
1K7B
 
 | | NMR Solution Structure of sTva47, the Viral-Binding Domain of Tva | | Descriptor: | SUBGROUP A ROUS SARCOMA VIRUS RECEPTOR PG800 AND PG950 | | Authors: | Tonelli, M, Peters, R.J, James, T.L, Agard, D.A. | | Deposit date: | 2001-10-18 | | Release date: | 2001-12-19 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the viral binding domain of Tva, the cellular receptor for subgroup A avian leukosis and sarcoma virus.
FEBS Lett., 509, 2001
|
|
1K81
 
 | |
1K8B
 
 | |
