7A64
 
 | |
7AAF
 
 | |
7AAO
 
 | |
7AC1
 
 | | Solution structure of the TAF4-RST domain | | Descriptor: | Transcription initiation factor TFIID subunit 4 | | Authors: | Staby, L, Davidsen, R, Bugge, K, Skriver, K, Kragelund, B.B. | | Deposit date: | 2020-09-09 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | alpha alpha-hub coregulator structure and flexibility determine transcription factor binding and selection in regulatory interactomes.
J.Biol.Chem., 298, 2022
|
|
7ACB
 
 | |
7ACE
 
 | |
7ACS
 
 | |
7ACT
 
 | |
7AEP
 
 | | Solution structure of U1-A RRM2 (190-282) | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Campagne, S, Allain, F.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands.
Nucleic Acids Res., 49, 2021
|
|
7AFQ
 
 | | Ribosome binding factor A (RbfA) | | Descriptor: | Ribosome-binding factor A | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2020-12-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7AFR
 
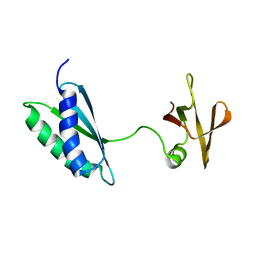 | | Ribosome maturation factor RimP (apo) | | Descriptor: | Ribosome maturation factor RimP | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7ALD
 
 | |
7ALU
 
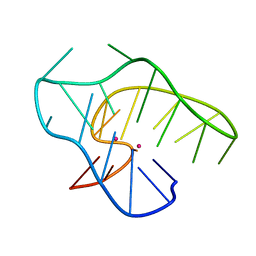 | |
7AQT
 
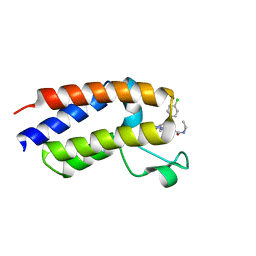 | | NMR2 structure of BRD4-BD2 in complex with iBET-762 | | Descriptor: | 2-[(4S)-6-(4-chlorophenyl)-8-methoxy-1-methyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-N-ethylacetamide, Bromodomain-containing protein 4 | | Authors: | Orts, J, Torres, F, Milbradt, A.G, Walser, R. | | Deposit date: | 2020-10-23 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | NMR Molecular Replacement Provides New Insights into Binding Modes to Bromodomains of BRD4 and TRIM24.
J.Med.Chem., 65, 2022
|
|
7ASY
 
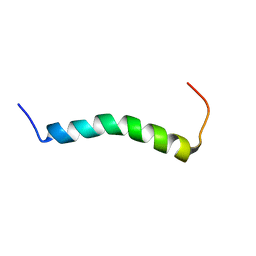 | |
7AT7
 
 | |
7ATB
 
 | |
7ATY
 
 | | Solution NMR structure of the SH3 domain of human Caskin1 | | Descriptor: | Caskin-1 | | Authors: | Toke, O, Koprivanacz, K, Radnai, L, Mero, B, Juhasz, T, Liliom, K, Buday, L. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the SH3 Domain of Human Caskin1 Validates the Lack of a Typical Peptide Binding Groove and Supports a Role in Lipid Mediator Binding.
Cells, 10, 2021
|
|
7ATZ
 
 | |
7AVA
 
 | |
7AVB
 
 | |
7AY8
 
 | | NMR solution structure of Tbo-IT2 | | Descriptor: | Tbo-IT2 | | Authors: | Mineev, K.S, Kornilov, F.D, Lushpa, V.A, Korolkova, Y.A, Maleeva, E.E, Andreev, Y.A, Kozlov, S.A, Arseniev, A.S. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insectotoxin from Tibellus Oblongus Spider Venom Presents Novel Adaptation of ICK Fold.
Toxins, 13, 2021
|
|
7B2B
 
 | | Solution structure of a non-covalent extended docking domain complex of the Pax NRPS: PaxA T1-CDD/PaxB NDD | | Descriptor: | Amino acid adenylation domain-containing protein, Peptide synthetase PaxA | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-16 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B2F
 
 | | Solution structure of the Pax NRPS docking domain PaxB NDD | | Descriptor: | Peptide synthetase XpsB (Modular protein) | | Authors: | Watzel, J, Sarawi, S, Duchardt-Ferner, E, Bode, H.B, Woehnert, J. | | Deposit date: | 2020-11-26 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cooperation between a T Domain and a Minimal C-Terminal Docking Domain to Enable Specific Assembly in a Multiprotein NRPS.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7B3J
 
 | | Dynamic complex between all-D-enantiomeric peptide D3 with wild-type amyloid precursor protein 672-726 fragment (amyloid beta 1-55) | | Descriptor: | D3 all D-enantimeric peptide, Isoform L-APP677 of Amyloid-beta precursor protein | | Authors: | Bocharov, E.V, Volynsky, P.E, Okhrimenko, I.S, Urban, A.S. | | Deposit date: | 2020-12-01 | | Release date: | 2021-01-13 | | Last modified: | 2021-12-08 | | Method: | SOLUTION NMR | | Cite: | All - d - Enantiomeric Peptide D3 Designed for Alzheimer's Disease Treatment Dynamically Interacts with Membrane-Bound Amyloid-beta Precursors.
J.Med.Chem., 64, 2021
|
|
