6S10
 
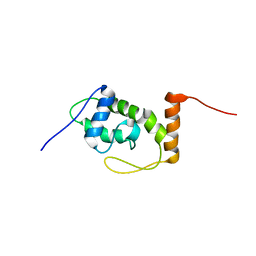 | |
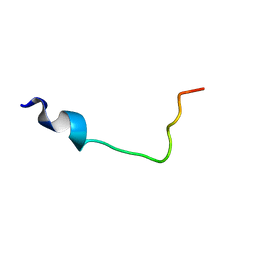
6S2D
 
 | |
6S3W
 
 | | Solution NMR Structure of TolAIII Bound to a Peptide Derived from the N-terminus of TolB | | Descriptor: | Cell envelope integrity/translocation protein TolA, TolBp | | Authors: | Kleanthous, C, Redfield, C, Rajasekar, K, Holmes, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | The lipoprotein Pal stabilises the bacterial outer membrane during constriction by a mobilisation-and-capture mechanism.
Nat Commun, 11, 2020
|
|
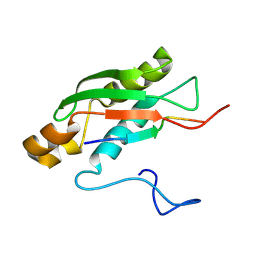
6SAA
 
 | | M-TRTX-Preg1a (Poecilotheria regalis) | | Descriptor: | U1-theraphotoxin-Pf3 | | Authors: | Meudal, H, Landon, C, Delmas, A.F. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists.
J.Med.Chem., 63, 2020
|
|
6SAB
 
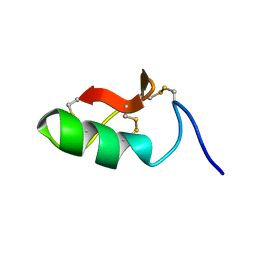 | | M-BUTX-Ptr1a (Parabuthus transvaalicus) | | Descriptor: | M-BUTX-Ptr1a | | Authors: | Meudal, H, Landon, C, Delmas, A.F. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists.
J.Med.Chem., 63, 2020
|
|
6SAI
 
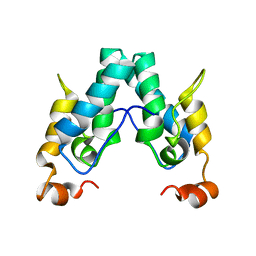 | |
6SAP
 
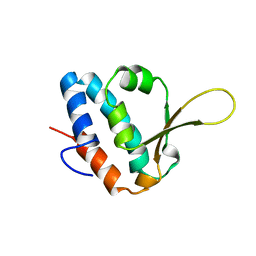 | |
6SCW
 
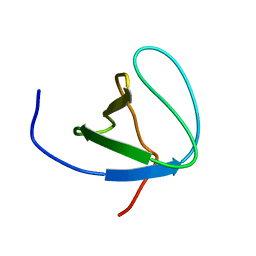 | |
6SDW
 
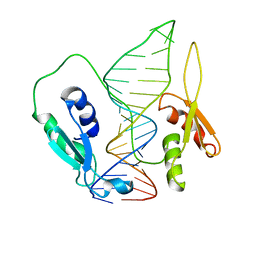 | |
6SDY
 
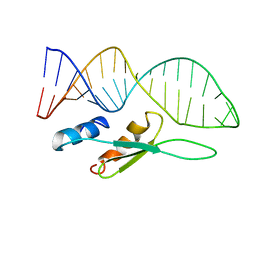 | |
6SFT
 
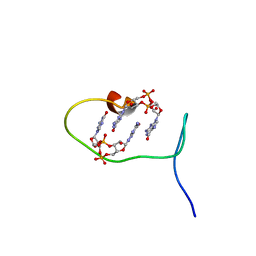 | | Solution structure of protein ARR_CleD in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Two-component receiver protein CleD | | Authors: | Habazettl, J, Hee, C.S, Jenal, U, Schirmer, T, Grzesiek, S. | | Deposit date: | 2019-08-02 | | Release date: | 2020-06-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Intercepting second-messenger signaling by rationally designed peptides sequestering c-di-GMP.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SGO
 
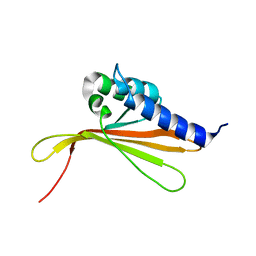 | | NMR structure of MLP124017 | | Descriptor: | Secreted protein | | Authors: | Barthe, P, de Guillen, K, Padilla, A, Hecker, A. | | Deposit date: | 2019-08-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural genomics applied to the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine knot and NTF2-like protein folds.
Sci Rep, 9, 2019
|
|
6SJW
 
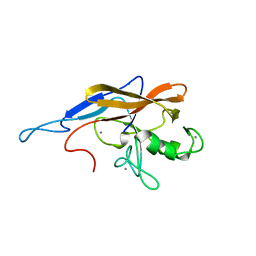 | | Structure of the self-processing module of iron-regulated FrpC of N. Meningitidis with calcium ions | | Descriptor: | CALCIUM ION, Iron-regulated protein FrpC | | Authors: | Kuban, V, Macek, P, Hritz, J, Nechvatalova, K, Nedbalcova, K, Faldyna, M, Zidek, L, Bumba, L. | | Deposit date: | 2019-08-14 | | Release date: | 2020-02-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Ca 2+ -Dependent Self-Processing Activity of Repeat-in-Toxin Proteins.
Mbio, 11, 2020
|
|
6SJX
 
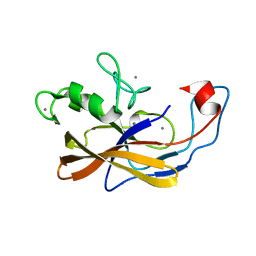 | | Precursor structure of the self-processing module of iron-regulated FrpC of N. Meningitidis with calcium ions | | Descriptor: | CALCIUM ION, Iron-regulated protein FrpC | | Authors: | Kuban, V, Macek, P, Hritz, J, Nechvatalova, K, Nedbalcova, K, Faldyna, M, Zidek, L, Bumba, L. | | Deposit date: | 2019-08-14 | | Release date: | 2020-02-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Ca 2+ -Dependent Self-Processing Activity of Repeat-in-Toxin Proteins.
Mbio, 11, 2020
|
|
6SLY
 
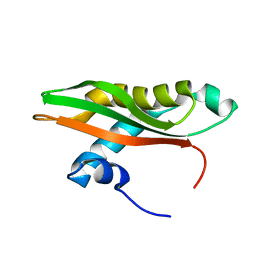 | |
6SNJ
 
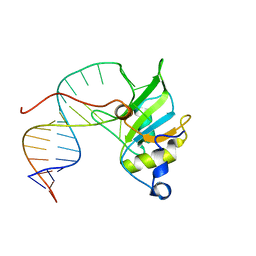 | |
6SO0
 
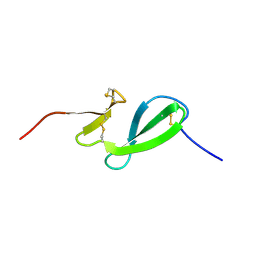 | | NMR solution structure of the family 14 carbohydrate binding module (CBM14) from human chitotriosidase | | Descriptor: | Chitotriosidase-1 | | Authors: | Madland, E, Crasson, O, Vandevenne, M, Sorlie, M, Aachmann, F.L. | | Deposit date: | 2019-08-28 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR and Fluorescence Spectroscopies Reveal the Preorganized Binding Site in Family 14 Carbohydrate-Binding Module from Human Chitotriosidase.
Acs Omega, 4, 2019
|
|
6SO9
 
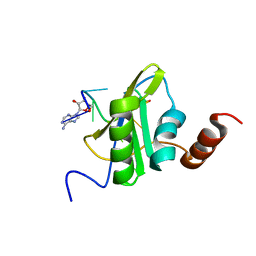 | |
6SOE
 
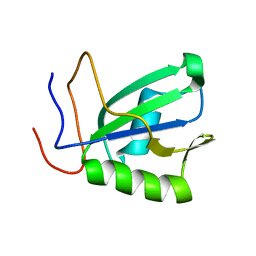 | | Mouse RBM20 RRM domain | | Descriptor: | RNA-binding protein 20 | | Authors: | Mackereth, C.D, Upadhyay, S.K. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis of UCUU RNA motif recognition by splicing factor RBM20.
Nucleic Acids Res., 48, 2020
|
|
6SOW
 
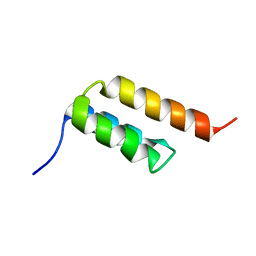 | |
6SUU
 
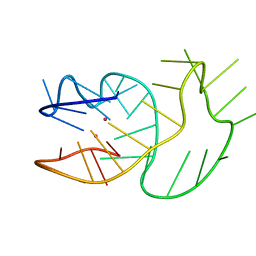 | |
6SVC
 
 | | Protein allostery of the WW domain at atomic resolution: apo structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SVE
 
 | | Protein allostery of the WW domain at atomic resolution: pCdc25C bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-10-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SVH
 
 | | Protein allostery of the WW domain at atomic resolution: FFpSPR bound structure | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Strotz, D, Orts, J, Friedmann, M, Guntert, P, Vogeli, B, Riek, R. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-30 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein Allostery at Atomic Resolution.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6SX3
 
 | | Intercalation of heterocyclic ligand between quartets in G-rich tetrahelical structure | | Descriptor: | VK2, ~{N}2,~{N}6-bis(1-methylquinolin-1-ium-3-yl)pyridine-2,6-dicarboxamide | | Authors: | Kotar, A, Kocman, V, Plavec, J. | | Deposit date: | 2019-09-24 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Intercalation of a Heterocyclic Ligand between Quartets in a G-Rich Tetrahelical Structure.
Chemistry, 26, 2020
|
|
