5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
5LMY
 
 | |
5LNF
 
 | | Solution NMR structure of farnesylated PEX19, C-terminal domain | | Descriptor: | FARNESYL, Peroxisomal biogenesis factor 19 | | Authors: | Emmanouilidis, L, Schuetz, U, Tripsianes, K, Madl, T, Radke, J, Rucktaeschel, R, Wilmanns, M, Schliebs, W, Erdmann, R, Sattler, M. | | Deposit date: | 2016-08-04 | | Release date: | 2017-03-15 | | Last modified: | 2019-09-11 | | Method: | SOLUTION NMR | | Cite: | Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19.
Nat Commun, 8, 2017
|
|
5LO2
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaTyr | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO3
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPaOMe | | Authors: | Baker, E.G, Hudson, K.L, Williams, C, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LO4
 
 | | Engineering protein stability with atomic precision in a monomeric miniprotein | | Descriptor: | PPa-CH3 | | Authors: | Baker, E.G, Williams, C, Hudson, K.L, Bartlett, G.G, Heal, J.W, Sessions, R.B, Crump, M.P, Woolfson, D.N. | | Deposit date: | 2016-08-08 | | Release date: | 2017-05-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering protein stability with atomic precision in a monomeric miniprotein.
Nat. Chem. Biol., 13, 2017
|
|
5LQG
 
 | |
5LQH
 
 | |
5LQV
 
 | | Spatial structure of the lentil lipid transfer protein in complex with anionic lysolipid LPPG | | Descriptor: | 1-MYRISTOYL-2-HYDROXY-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL)], Non-specific lipid-transfer protein 2 | | Authors: | Mineev, K.S, Shenkarev, Z.O, Arseniev, A.S, Melnikova, D.N, Finkina, E.I, Ovchinnikova, T.V. | | Deposit date: | 2016-08-17 | | Release date: | 2017-06-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Ligand Binding Properties of the Lentil Lipid Transfer Protein: Molecular Insight into the Possible Mechanism of Lipid Uptake.
Biochemistry, 56, 2017
|
|
5LSD
 
 | | recombinant mouse Nerve Growth Factor | | Descriptor: | Beta-nerve growth factor | | Authors: | Paoletti, F, de Chiara, C, Kelly, G, Lamba, D, Cattaneo, A, Pastore, A. | | Deposit date: | 2016-08-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Conformational Rigidity within Plasticity Promotes Differential Target Recognition of Nerve Growth Factor.
Front Mol Biosci, 3, 2016
|
|
5LSN
 
 | |
5LUE
 
 | | Minor form of the recombinant cytotoxin-1 from N. oxiana | | Descriptor: | VC-1=CYTOTOXIN | | Authors: | Dubovskii, P.V, Dubinnyi, M.A, Shulepko, M.A, Lyukmanova, E.N, Dolgikh, D.A, Kirpichnikov, M.P, Efremov, R.G. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic "Portraits" of Recombinant and Native Cytotoxin I from Naja oxiana: How Close Are They?
Biochemistry, 56, 2017
|
|
5LV6
 
 | | N-terminal motif dimerization of EGFR transmembrane domain in bicellar environment | | Descriptor: | Epidermal growth factor receptor | | Authors: | Bragin, P, Bocharov, E, Mineev, K, Bocharova, O, Arseniev, A. | | Deposit date: | 2016-09-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Conformation of the Epidermal Growth Factor Receptor Transmembrane Domain Dimer Dynamically Adapts to the Local Membrane Environment.
Biochemistry, 56, 2017
|
|
5LVF
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Thr4 phosphorylated CTD peptide | | Descriptor: | PRO-SER-TYR-SER-PRO-PTH-SER-PRO-SER-TYR-SER-PRO-THR-SER-PRO-SER, Regulator of Ty1 transposition protein 103 | | Authors: | Jasnovidova, O, Kubicek, K, Krejcikova, M, Stefl, R. | | Deposit date: | 2016-09-14 | | Release date: | 2017-05-10 | | Last modified: | 2019-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural insight into recognition of phosphorylated threonine-4 of RNA polymerase II C-terminal domain by Rtt103p.
EMBO Rep., 18, 2017
|
|
5LVY
 
 | |
5LW4
 
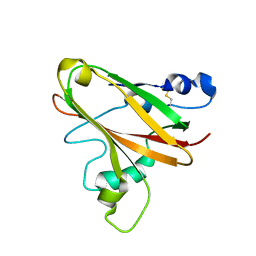 | |
5LW8
 
 | |
5LWC
 
 | | NMR solution structure of bacteriocin BacSp222 from Staphylococcus pseudintermedius 222 | | Descriptor: | Bacteriocin BacSp222 | | Authors: | Nowakowski, M.E, Ejchart, A.O, Jaremko, L, Wladyka, B, Mak, P. | | Deposit date: | 2016-09-15 | | Release date: | 2017-10-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Spatial attributes of the four-helix bundle group of bacteriocins - The high-resolution structure of BacSp222 in solution.
Int.J.Biol.Macromol., 107, 2018
|
|
5LWJ
 
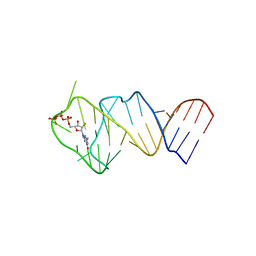 | | Solution NMR structure of the GTP binding Class II RNA aptamer-ligand-complex containing a protonated adenine nucleotide with a highly shifted pKa. | | Descriptor: | GTP Class II RNA (34-MER), GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Wolter, A.C, Weickhmann, A.K, Nasiri, A.H, Hantke, K, Ohlenschlaeger, O, Wunderlich, C.H, Kreutz, C, Duchardt-Ferner, E, Woehnert, J. | | Deposit date: | 2016-09-17 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Stably Protonated Adenine Nucleotide with a Highly Shifted pKa Value Stabilizes the Tertiary Structure of a GTP-Binding RNA Aptamer.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LXJ
 
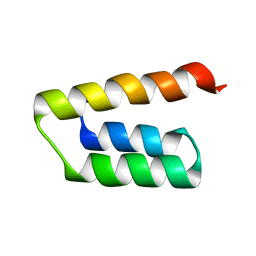 | |
5LXK
 
 | | NMR structure of the C-terminal domain of the Bacteriophage T5 decoration protein pb10. | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5LXL
 
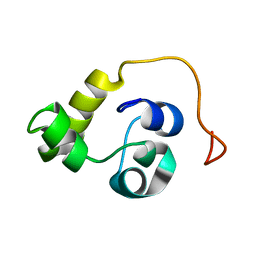 | | NMR structure of the N-terminal domain of the Bacteriophage T5 decoration protein pb10 | | Descriptor: | Decoration protein | | Authors: | Vernhes, E, Gilquin, B, Cuniasse, P, Boulanger, P, Zinn-Justin, S. | | Deposit date: | 2016-09-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | High affinity anchoring of the decoration protein pb10 onto the bacteriophage T5 capsid.
Sci Rep, 7, 2017
|
|
5M0A
 
 | |
5M1G
 
 | |
5M1H
 
 | |
