2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RS7
 
 | | Solution structure of the second dsRBD from RNA helicase A | | Descriptor: | ATP-dependent RNA helicase A | | Authors: | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RS8
 
 | | Solution structure of the N-terminal RNA recognition motif of NonO | | Descriptor: | Non-POU domain-containing octamer-binding protein | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA recognition motif of NonO
To be Published
|
|
2RS9
 
 | | Solution structure of the bromodomain of human BRPF1 in complex with histone H4K5ac peptide | | Descriptor: | Acetylated lysine 5 of peptide from Histone H4, Peregrin | | Authors: | Qin, X, Nagashima, T, Umehara, T, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Site-specific histone recognition by the bromodomain of Brpf1 and the role in MOZ/MORF histone acetyltransferase complexes
To be Published
|
|
2RSC
 
 | |
2RSD
 
 | | Solution structure of the plant homeodomain (PHD) of the E3 SUMO ligase Siz1 from rice | | Descriptor: | E3 SUMO-protein ligase SIZ1, ZINC ION | | Authors: | Shindo, H, Tsuchiya, W, Suzuki, R, Yamazaki, T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PHD finger of the SUMO ligase Siz/PIAS family in rice reveals specific binding for methylated histone H3 at lysine 4 and arginine 2
Febs Lett., 586, 2012
|
|
2RSE
 
 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
2RSF
 
 | | Complex structure of WWE in RNF146 with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, E3 ubiquitin-protein ligase RNF146 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-01-31 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Complex structure of WWE domain in RNF146 with ATP
To be Published
|
|
2RSG
 
 | | Solution structure of the CERT PH domain | | Descriptor: | Collagen type IV alpha-3-binding protein | | Authors: | Sugiki, T, Takeuchi, K, Tokunaga, Y, Kumagai, K, Kawano, M, Nishijima, M, Hanada, K, Takahashi, H, Shimada, I. | | Deposit date: | 2012-02-25 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the Golgi association by the pleckstrin homology domain of the ceramide trafficking protein (CERT)
J.Biol.Chem., 287, 2012
|
|
2RSH
 
 | |
2RSI
 
 | |
2RSJ
 
 | |
2RSK
 
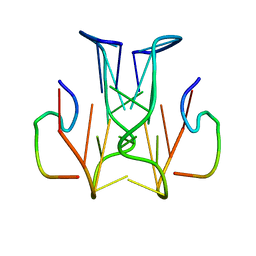 | | RNA aptamer against prion protein in complex with the partial binding peptide | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3'), partial binding peptide of Major prion protein | | Authors: | Mashima, T, Nishikawa, F, Kamatari, Y.O, Fujiwara, H, Nishikawa, S, Kuwata, K, Katahira, M. | | Deposit date: | 2012-03-08 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Anti-prion activity of an RNA aptamer and its structural basis
Nucleic Acids Res., 41, 2013
|
|
2RSM
 
 | | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65 (ICT2) | | Descriptor: | Probable peptide chain release factor C12orf65 homolog, mitochondrial | | Authors: | Enomoto, M, Tochio, N, Tomizawa, T, Koshiba, S, Guntert, P, Kigawa, T, Yokoyama, S, Nameki, N. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and siRNA-mediated knockdown analysis of the mitochondrial disease-related protein C12orf65.
Proteins, 2012
|
|
2RSN
 
 | |
2RSO
 
 | |
2RSQ
 
 | | Copper(I) loaded form of the first domain of the human copper chaperone for SOD1, CCS | | Descriptor: | COPPER (I) ION, Copper chaperone for superoxide dismutase | | Authors: | Banci, L, Bertini, I, Cantini, F, Kozyreva, T, Rubino, J.T. | | Deposit date: | 2012-05-15 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Copper(I) loaded form of the first domain of the human copper chaperone for SOD1, CCS
To be Published
|
|
2RST
 
 | | NMR structure of the C-terminal domain of EW29 | | Descriptor: | 29-kDa galactose-binding lectin | | Authors: | Hemmi, H. | | Deposit date: | 2012-05-29 | | Release date: | 2013-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the C-terminal domain of R-type lectin from the earthworm Lumbricus terrestris
Febs J., 280, 2013
|
|
2RSU
 
 | | Alternative structure of Ubiquitin | | Descriptor: | Ubiquitin | | Authors: | Kitazawa, S, Kameda, T, Yagi-Utsumi, M, Kato, K, Kitahara, R. | | Deposit date: | 2012-06-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Q41N Variant of Ubiquitin as a Model for the Alternatively Folded N2 State of Ubiquitin
Biochemistry, 52, 2013
|
|
2RSV
 
 | |
2RSW
 
 | |
2RSX
 
 | | Solution structure of IseA, an inhibitor protein of DL-endopeptidases from Bacillus subtilis | | Descriptor: | Uncharacterized protein yoeB | | Authors: | Arai, R, Li, H, Tochio, N, Fukui, S, Kobayashi, N, Kitaura, C, Watanabe, S, Kigawa, T, Sekiguchi, J. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of IseA, an Inhibitor Protein of DL-Endopeptidases from Bacillus subtilis, Reveals a Novel Fold with a Characteristic Inhibitory Loop
J.Biol.Chem., 287, 2012
|
|
2RSY
 
 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
2RT3
 
 | | Solution structure of the second RRM domain of Nrd1 | | Descriptor: | Negative regulator of differentiation 1 | | Authors: | Kobayashi, A, Kanaba, T, Mishima, M. | | Deposit date: | 2013-04-16 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the second RRM domain of Nrd1, a fission yeast MAPK target RNA binding protein, and implication for its RNA recognition and regulation
Biochem.Biophys.Res.Commun., 437, 2013
|
|
2RT4
 
 | |
