9MYZ
 
 | |
9MZ0
 
 | |
9MZ1
 
 | | Structure of human endothelial nitric oxide synthase heme domain bound with 7-(3-aminomethyl)phenyl-6-fluoro-4-methylquinoin-2-amine | | Descriptor: | (7P)-7-[3-(aminomethyl)phenyl]-6-fluoro-4-methylquinolin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2025-01-22 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and synthesis of 7-aryl-6-fluoro-2-aminoquinolines as potent and highly selective human neuronal nitric oxide synthase inhibitors
To Be Published
|
|
9MZ2
 
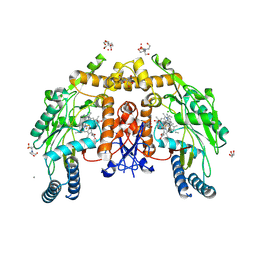 | | Structure of human endothelial nitric oxide synthase heme domain bound with 2-(2-amino-6-fluoro-4-methylquinolin-7-yl)-5-(aminomethyl)phenol | | Descriptor: | (2M)-2-(2-amino-6-fluoro-4-methylquinolin-7-yl)-5-(aminomethyl)phenol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2025-01-22 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Design and synthesis of 7-aryl-6-fluoro-2-aminoquinolines as potent and highly selective human neuronal nitric oxide synthase inhibitors
To Be Published
|
|
9MZ3
 
 | | Structure of human endothelial nitric oxide synthase heme domain bound 2-(2-amino-6-fluoro-4-methylquinolin-7-yl)-5-(2-aminoethyl)phenol | | Descriptor: | (2M)-5-(2-aminoethyl)-2-(2-amino-6-fluoro-4-methylquinolin-7-yl)phenol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2025-01-22 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and synthesis of 7-aryl-6-fluoro-2-aminoquinolines as potent and highly selective human neuronal nitric oxide synthase inhibitors
To Be Published
|
|
9MZ4
 
 | | Structure of human endothelial nitric oxide synthase heme domain bound 7-(3-(aminomethyl)-4-(cyclopropylmethoxy)phenyl)-6-fluoro-4-methylquinolin-2-amine | | Descriptor: | (7P)-7-[3-(aminomethyl)-4-(cyclopropylmethoxy)phenyl]-6-fluoro-4-methylquinolin-2-amine, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2025-01-22 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design and synthesis of 7-aryl-6-fluoro-2-aminoquinolines as potent and highly selective human neuronal nitric oxide synthase inhibitors
To Be Published
|
|
9N5P
 
 | | CRYSTAL STRUCTURE OF HUMAN IGG2 FC FRAGMENT-FC-GAMMA RECEPTOR IIA COMPLEX H131 VARIANT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 1 of Immunoglobulin heavy constant gamma 2, ... | | Authors: | Tolbert, W.D, Pazgier, M. | | Deposit date: | 2025-02-04 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Cross-species analysis of FcgRIIa/b (CD32a/b) polymorphisms at position 131: structural and functional insights into the mechanism of IgG- mediated phagocytosis in human and macaque.
Front Immunol, 2025
|
|
9NAZ
 
 | | Structure of SARS-CoV-2 NSP14 bound to N-((4-vinylthiazol-2-yl)methyl)-1H-pyrazole-3-carboxamide | | Descriptor: | Guanine-N7 methyltransferase nsp14, N-[(4-ethenyl-1,3-thiazol-2-yl)methyl]-1H-pyrazole-3-carboxamide, PHOSPHATE ION, ... | | Authors: | Coker, J.A, Sun, R, Polzer, P.M, Jung, J, Stauffer, S.R. | | Deposit date: | 2025-02-13 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Development of NSP14 Exonuclease Inhibitors Confounded by Batch-to-Batch Variability
Biorxiv, 2025
|
|
9NFP
 
 | | Structure of SARS-CoV-2 NSP14 bound to N-((4-cyclopropylthiazol-2-yl)methyl)-1H-pyrazole-3-carboxamide | | Descriptor: | Guanine-N7 methyltransferase nsp14, N-[(4-cyclopropyl-1,3-thiazol-2-yl)methyl]-1H-pyrazole-3-carboxamide, PHOSPHATE ION, ... | | Authors: | Coker, J.A, Sun, R, Polzer, P.M, Jung, J, Stauffer, S.R. | | Deposit date: | 2025-02-21 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Development of NSP14 Exonuclease Inhibitors Confounded by Batch-to-Batch Variability
Biorxiv, 2025
|
|
9NHA
 
 | | Structure of SARS-CoV-2 NSP14 bound to N-((4-isopropylthiazol-2-yl)methyl)-1H-pyrazole-3-carboxamide | | Descriptor: | Guanine-N7 methyltransferase nsp14, N-{[4-(propan-2-yl)-1,3-thiazol-2-yl]methyl}-1H-pyrazole-3-carboxamide, PHOSPHATE ION, ... | | Authors: | Coker, J.A, Sun, R, Polzer, P.M, Jung, J, Stauffer, S.R. | | Deposit date: | 2025-02-24 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Development of NSP14 Exonuclease Inhibitors Confounded by Batch-to-Batch Variability
Biorxiv, 2025
|
|
9NHU
 
 | | Structure of SARS-CoV-2 NSP14 bound to 5-(((cyclopropylmethyl)amino)methyl)-N-((4-cyclopropylthiazol-2-yl)methyl)-1H-pyrazole-3-carboxamide | | Descriptor: | 5-{[(cyclopropylmethyl)amino]methyl}-N-[(4-cyclopropyl-1,3-thiazol-2-yl)methyl]-1H-pyrazole-3-carboxamide, Guanine-N7 methyltransferase nsp14, PHOSPHATE ION, ... | | Authors: | Coker, J.A, Sun, R, Polzer, P.M, Jung, J, Stauffer, S.R. | | Deposit date: | 2025-02-25 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Development of NSP14 Exonuclease Inhibitors Confounded by Batch-to-Batch Variability
Biorxiv, 2025
|
|
9NIO
 
 | | SARS-CoV-2 NSP14 bound to N-((2-ethynylthiazol-4-yl)methyl)-1H-pyrazole-3-carboxamide | | Descriptor: | Guanine-N7 methyltransferase nsp14, N-[(2-ethynyl-1,3-thiazol-4-yl)methyl]-1H-pyrazole-3-carboxamide, PHOSPHATE ION, ... | | Authors: | Coker, J.A, Sun, R, Polzer, P.M, Jung, J, Stauffer, S.R. | | Deposit date: | 2025-02-26 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-Based Development of NSP14 Exonuclease Inhibitors Confounded by Batch-to-Batch Variability
Biorxiv, 2025
|
|
9NJG
 
 | | Structure of SARS-CoV-2 NSP14 bound to N-((4-cyclopropylthiazol-2-yl)methyl)-1H-pyrazolo[3,4-b]pyridine-3-carboxamide | | Descriptor: | Guanine-N7 methyltransferase nsp14, N-[(4-cyclopropyl-1,3-thiazol-2-yl)methyl]-1H-pyrazolo[3,4-b]pyridine-3-carboxamide, PHOSPHATE ION, ... | | Authors: | Coker, J.A, Sun, R, Polzer, P.M, Jung, J, Stauffer, S.R. | | Deposit date: | 2025-02-27 | | Release date: | 2025-12-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-Based Development of NSP14 Exonuclease Inhibitors Confounded by Batch-to-Batch Variability
Biorxiv, 2025
|
|
9NK6
 
 | |
9NK7
 
 | |
9NK8
 
 | |
9NRX
 
 | |
9NRY
 
 | |
9NRZ
 
 | |
9OFO
 
 | | HCoV-229E S2P bound by three DH1533 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1533 FAB heavy chain, ... | | Authors: | Wrapp, D. | | Deposit date: | 2025-04-30 | | Release date: | 2025-12-03 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A potently neutralizing human antibody induces movement of the HCoV-229E receptor binding domain
To Be Published
|
|
9OFP
 
 | | HCoV-229E S2P bound by two DH1533 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1533 heavy chain, ... | | Authors: | Wrapp, D. | | Deposit date: | 2025-04-30 | | Release date: | 2025-12-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | A potently neutralizing human antibody induces movement of the HCoV-229E receptor binding domain
To Be Published
|
|
9OFQ
 
 | | HCoV-229E S2P bound by one DH1533 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1533 FAB heavy chain, ... | | Authors: | Wrapp, D. | | Deposit date: | 2025-04-30 | | Release date: | 2025-12-03 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A potently neutralizing human antibody induces movement of the HCoV-229E receptor binding domain
To Be Published
|
|
9OIC
 
 | | Structure of shaker-IR-I384R | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | Authors: | Liu, Y, Contreras, G.F. | | Deposit date: | 2025-05-06 | | Release date: | 2025-12-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Closed state structure of the pore revealed by uncoupled Shaker K + channel.
Nat Commun, 16, 2025
|
|
9OL5
 
 | |
9OL6
 
 | |
