8Q4D
 
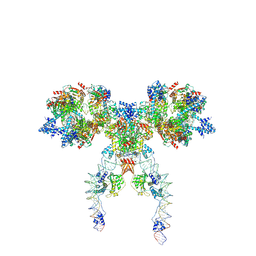 | | IstA-IstB(E167Q) Strand Transfer Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA (118-MER) / TIR-transferred strand, ... | | Authors: | de la Gandara, A, Spinola-Amilibia, M, Araujo-Bazan, L, Nunez-Ramirez, R, Berger, J.M, Arias-Palomo, E. | | Deposit date: | 2023-08-06 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Molecular basis for transposase activation by a dedicated AAA+ ATPase.
Nature, 630, 2024
|
|
8Q4F
 
 | | Structure of arbekacin bound Escherichia coli 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L16, ... | | Authors: | Majumdar, S, Parajuli, N.P, Ge, X, Emmerich, A, Sanyal, S. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of arbekacin bound Escherichia coli 70S ribosome
To be published
|
|
8Q4K
 
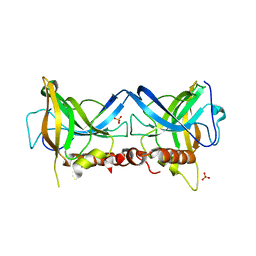 | | Crystal structure of Borrelia burgdorferi BB0158 | | Descriptor: | S2 lipoprotein, SULFATE ION | | Authors: | Brangulis, K, Tars, K. | | Deposit date: | 2023-08-07 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of chromosomally encoded outer surface lipoprotein BB0158 from Borrelia burgdorferi sensu stricto.
Ticks Tick Borne Dis, 15, 2024
|
|
8Q54
 
 | |
8Q5I
 
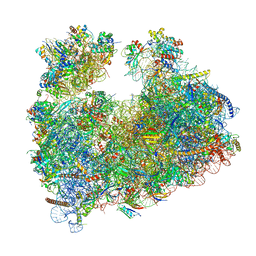 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | Descriptor: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
8Q62
 
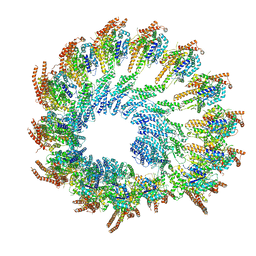 | | Early closed conformation of the g-tubulin ring complex | | Descriptor: | Gamma-tubulin complex component 2, Gamma-tubulin complex component 3, Gamma-tubulin complex component 4, ... | | Authors: | Llorca, O, Serna, M, Fernandez-Leiro, R. | | Deposit date: | 2023-08-10 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Transition of human gamma-tubulin ring complex into a closed conformation during microtubule nucleation.
Science, 383, 2024
|
|
8Q6J
 
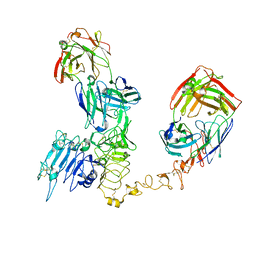 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
8Q6L
 
 | | human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, Carbonic anhydrase 1, SODIUM ION, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-08-13 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol
To Be Published
|
|
8Q6T
 
 | |
8Q78
 
 | | Structure of the FP specific VHH TPP-3077 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, TPP-3077 VHH | | Authors: | Andersen, G.R, Pedersen, D.V. | | Deposit date: | 2023-08-15 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.225 Å) | | Cite: | Characterization of the bispecific VHH antibody tarperprumig (ALXN1820) specific for properdin and designed for low-volume administration.
Mabs, 16, 2024
|
|
8Q7G
 
 | | Human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol pH 7.0 | | Descriptor: | 1,1-bis(oxidanyl)-3,4-dihydro-2,1$l^{4}-benzoxaborinine, ACETATE ION, Carbonic anhydrase 1, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Human Carbonic Anhydrase I in complex with 3,4-dihydro-1H-benzo[c][1,2]oxaborinin-1-ol pH 7.0
To Be Published
|
|
8Q7X
 
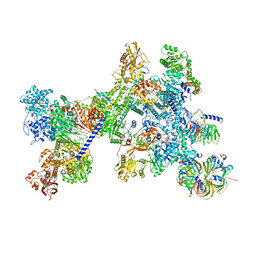 | | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 4) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7Z
 
 | |
8Q84
 
 | |
8Q87
 
 | |
8QBT
 
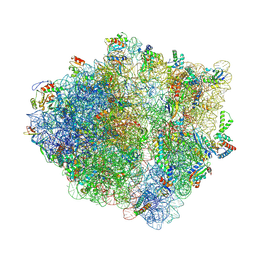 | | E. coli ApdP-stalled ribosomal complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Morici, M, Wilson, D.N. | | Deposit date: | 2023-08-25 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | RAPP-containing arrest peptides induce translational stalling by short circuiting the ribosomal peptidyltransferase activity.
Nat Commun, 15, 2024
|
|
8QBX
 
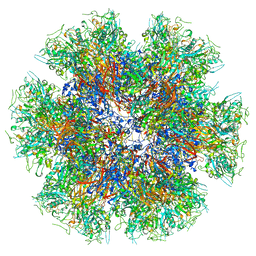 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8QC4
 
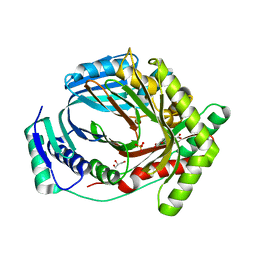 | | M. tuberculosis salicylate synthase MbtI in complex with 5-(3-carboxyphenyl)furan-2-carboxylic acid | | Descriptor: | 5-(3-carboxyphenyl)furan-2-carboxylic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Mori, M, Villa, S, Meneghetti, F, Bellinzoni, M. | | Deposit date: | 2023-08-25 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Structural Study of a New MbtI-Inhibitor Complex: Towards an Optimized Model for Structure-Based Drug Discovery.
Pharmaceuticals, 16, 2023
|
|
8QCQ
 
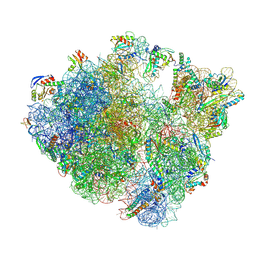 | | B. subtilis ApdA-stalled ribosomal complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Morici, M, Wilson, D.N. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | RAPP-containing arrest peptides induce translational stalling by short circuiting the ribosomal peptidyltransferase activity.
Nat Commun, 15, 2024
|
|
8QDO
 
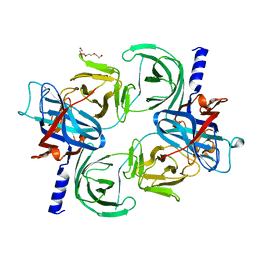 | | Crystal structure of the tegument protein UL82 (pp71) from Human Cytomegalovirus | | Descriptor: | Protein pp71, TETRAETHYLENE GLYCOL | | Authors: | Bresch, I.P, Eberhage, J, Reubold, T.F, Eschenburg, S. | | Deposit date: | 2023-08-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the tegument protein UL82 (pp71) from human cytomegalovirus.
Protein Sci., 33, 2024
|
|
8QF2
 
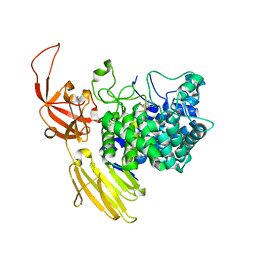 | | Beta-L-Arabinofurano-cyclitol Aziridines are Cysteine-directed Broad-spectrum Inhibitors and Activity-based Probes for Retaining Beta-L-arabinofuranosidases | | Descriptor: | (1~{S},2~{S},3~{S},4~{R})-4-azanyl-3-(hydroxymethyl)cyclopentane-1,2-diol, Non-reducing end beta-L-arabinofuranosidase, ZINC ION | | Authors: | Borlandelli, V, Offen, W.A, Moroz, O, Nin-Hill, A, McGregor, N, Binkhorst, L, Armstrong, Z, Ishiwata, A, Artola, M, Rovira, C, Davies, G.J, Overkleeft, H. | | Deposit date: | 2023-09-02 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | beta-l- Arabino furano-cyclitol Aziridines Are Covalent Broad-Spectrum Inhibitors and Activity-Based Probes for Retaining beta-l-Arabinofuranosidases.
Acs Chem.Biol., 18, 2023
|
|
8QF7
 
 | |
8QF9
 
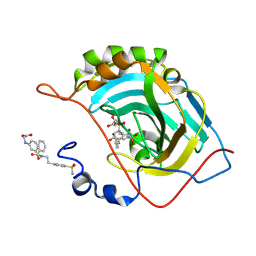 | |
8QFD
 
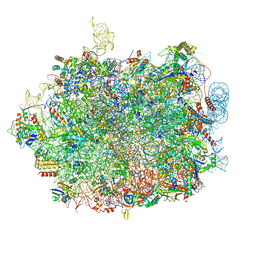 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Makhlouf, L, Kulathu, Y, Zeqiraj, E. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
8QFK
 
 | |






