8YTO
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTP
 
 | | Single-chain Fv antibody of E11 complex with NP-glycine under reducing conditions | | Descriptor: | 2-[2-(3-nitro-4-oxidanyl-phenyl)ethanoylamino]ethanoic acid, Single-chain Fv antibody of E11 | | Authors: | Yoshida, M, Hanazono, Y, Numoto, N, Ito, N, Oda, M. | | Deposit date: | 2024-03-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Affinity-matured antibody with a disulfide bond in H-CDR3 loop.
Arch.Biochem.Biophys., 758, 2024
|
|
8YTX
 
 | | Tubulin-RB3-TTL in complex with compound SI9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-~{N}-(4-methoxyphenyl)-~{N}-methyl-thieno[2,3-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Tubulin-RB3-TTL in complex with compound SI9
To Be Published
|
|
8YU9
 
 | | Tubulin-RB3-TTL in complex with compound SI10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloranylthieno[3,2-d]pyrimidin-4-yl)-7-methoxy-1,3-dihydroquinoxalin-2-one, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of tubulin-RB3-TTL in complex with compound AB10
To Be Published
|
|
8YUA
 
 | | Tubulin-RB3-TTL in complex with compound SI10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-~{N}-(4-methoxyphenyl)-~{N}-methyl-thieno[3,2-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The complex structure of tubulin-RB3-TTL in complex with compound SI10
To Be Published
|
|
8YUO
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 at the A-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 2024
|
|
8YUP
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and A4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-28 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 2024
|
|
8YUQ
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and dA4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 2024
|
|
8YUR
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and Am4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 2024
|
|
8YUS
 
 | | E. coli 70S ribosome complexed with P.putida tRNAIle2 and A(F)4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 2024
|
|
8YUT
 
 | | Cryo-EM structure of the amthamine-bound H2R-Gs complex | | Descriptor: | 5-(2-azanylethyl)-4-methyl-1,3-thiazol-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YUV
 
 | | Cryo-EM structure of the immepip-bound H3R-Gi complex | | Descriptor: | 4-(1H-imidazol-5-ylmethyl)piperidine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, Q, Tang, X, Wen, X, Cheng, S, Xiao, P, Zang, S, Shen, D, Jiang, L, Zheng, Y, Zhang, H, Xu, H, Mao, C, Zhang, M, Hu, W, Sun, J, Chen, Z, Zhang, Y. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs.
Adv Sci, 11, 2024
|
|
8YVG
 
 | | canine immunoproteasome 20S subunit in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type, Proteasome subunit beta, Proteasome subunit beta type-8, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-28 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
8YVP
 
 | | canine immunoproteasome 20S subunit in complex with compound 1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Kashima, A, Arai, Y. | | Deposit date: | 2024-03-29 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Optimization of alpha-amido boronic acids via cryo-electron microscopy analysis: Discovery of a novel highly selective immunoproteasome subunit LMP7 ( beta 5i)/LMP2 ( beta 1i) dual inhibitor.
Bioorg.Med.Chem., 109, 2024
|
|
8YYQ
 
 | | Structure of the HitB F328L mutant | | Descriptor: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(3-cyanophenyl)propanoyl]sulfamate | | Authors: | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2024-04-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 25, 2024
|
|
8YYR
 
 | | Structure of the HitB T293G mutant | | Descriptor: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(2-bromophenyl)propanoyl]sulfamate | | Authors: | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2024-04-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 25, 2024
|
|
8Z0K
 
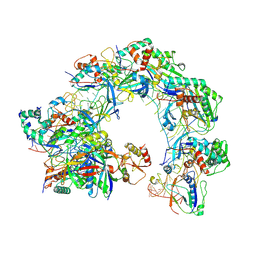 | | Cryo-EM structure of Cas8-HNH system at full R-loop state | | Descriptor: | DNA (37-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8Z1L
 
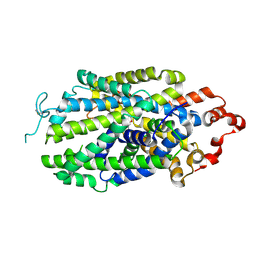 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom. | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8Z2H
 
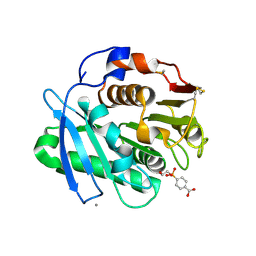 | | Substrate analog a010 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 4-[2-hydroxyethyloxy(oxidanyl)phosphoryl]benzoic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2I
 
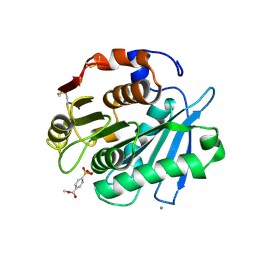 | | Substrate analog a011 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 2-hydroxyethyloxy-(4-methoxycarbonylphenyl)phosphinic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2J
 
 | | Substrate analog a012 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | (4-methoxycarbonylphenyl)-(2-phenylmethoxyethoxy)phosphinic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2K
 
 | | Substrate analog a013 bound form of PET-degrading cutinase mutant Cut190**SS_S176A | | Descriptor: | 4-[oxidanyl(2-phenylmethoxyethoxy)phosphoryl]benzoic acid, Alpha/beta hydrolase family protein, CALCIUM ION | | Authors: | Numoto, N, Kondo, F, Bekker, G.J, Liao, Z, Yamashita, M, Iida, A, Ito, N, Kamiya, N, Oda, M. | | Deposit date: | 2024-04-12 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dynamics of the Ca 2+ -regulated cutinase towards structure-based improvement of PET degradation activity.
Int.J.Biol.Macromol., 281, 2024
|
|
8Z2N
 
 | | Crystal structure of 5-phosphomethyl-2'-deoxyuridine (5-PmdU) glycinyltransferase gp46/PUGT from Pseudomonads phage PaMx11 in complex with dsDNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*AP*GP*TP*CP*GP*AP*CP*GP*AP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*GP*TP*CP*GP*AP*CP*TP*A)-3'), ... | | Authors: | Wen, Y, Guo, W.T, Wu, B.X. | | Deposit date: | 2024-04-13 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the biosynthetic mechanism of N alpha-GlyT and 5-NmdU hypermodifications of DNA.
Nucleic Acids Res., 52, 2024
|
|
8Z2W
 
 | | Crystal structure of apo- exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) | | Descriptor: | 2-methylpropylphosphonic acid, Glucan 1,3-beta-glucosidase A, SODIUM ION | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 2024
|
|
8Z3S
 
 | | Activation mechanism and novel binding site of the BKCa channel activator CTIBD | | Descriptor: | 4-[4-(4-chlorophenyl)-3-(trifluoromethyl)-1,2-oxazol-5-yl]benzene-1,3-diol, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Lee, N, Kim, S, Jo, H, Lee, N.Y, Jin, M.S, Park, C.S. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Activation mechanism and novel binding sites of the BK Ca channel activator CTIBD.
Life Sci Alliance, 7, 2024
|
|






